Frequency Band - Questions about ReHo and fALFF
Hi everybody, i've some questions about DPARSF and the frequency range which is selected for processing :
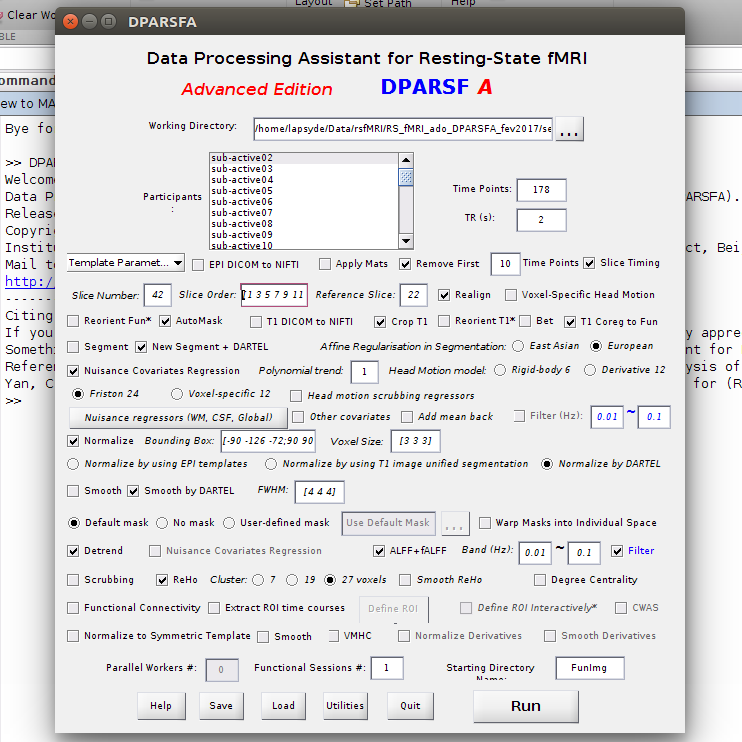
Q. 1 : If I want to chose a specific frequency band for ReHo analysis (like in this paper : doi:10.1371/
p.p1 {margin: 0.0px 0.0px 0.0px 0.0px; font: 7.0px Helvetica}journal.pone.0086818.: IMF1 : (0.1-0.25 Hz), IMF2 (0.1-0.04 Hz)...), is this filter has to be used (cf screenshot) ?