Error in Calculating Functional Connectivity by Seed based Correlation Anlyasis
DPARSFA error: array size
- Read more about DPARSFA error: array size
- 1 comment
- Log in or register to post comments
Dear all,
during the extraction of the ROI signals, I get the error message you can see in the picture. Apparently, DPARSFA is trying to create an array of size 279586x279586, or 582.4GB which is, needless to say, way too large for our computers. Is this a bug?
This error occurred when trying to perform nuisance regression on files that are already preprocessed (see pictures)
Cheers,
Sascha
Mask for Global Signal Regression
- Read more about Mask for Global Signal Regression
- 1 comment
- Log in or register to post comments
Dear all,
I want to perform Global Signal Regression on files which I have already preprocessed (swau) using another toolbox. For GSR, one can choose between "SPM apriori" and "Automask". This confuses me; What is the difference between the Option "Segment" for WM/CSF-regression and the option "Automask" for GSR? Also, can't I use the already existing brainmasks which I stored in the folder WorkingDirectory/Masks/WarpedMasks?
Thanks!
Cheers,
Sascha
Nomenclature of mask-files
- Read more about Nomenclature of mask-files
- 1 comment
- Log in or register to post comments
Dear all,
in the folder Masks/WarpedMasks, for each subject, there are three different files (now exemplary for subject 001):
- Sub_001_BrainMask_05_91x109x91.nii
- Sub_001_WhiteMask_09_91x109x91.nii
- Sub_001_CsfMask_07_91x109x91.nii
I wonder, what do the indices 05, 07 and 09 mean? Are they relevant for anything or is it just a compatibility question?
Thanks!
Cheers,
Sascha
with GSR and without GSR
- Read more about with GSR and without GSR
- 6 comments
- Log in or register to post comments
Dear DPARSFA experts,
Recently, I updated the DPARSF to the latest verison because of the new feture of GSR. In the analyis, I clicked the button "both with and without GSR". However, there is only one type of data after nuisance covariates regression. According to my understanding, there should be two types of data: one with GSR and another without GSR, and the results should have two. Furthermore, I did not know the results whether regress with global signal. Is anything wrong with my setting? Could you help me on this problem? Your repsonses will be appreciated.
About the mask for Alphasim correction
- Read more about About the mask for Alphasim correction
- 2 comments
- Log in or register to post comments
Dear professor Yan,
I have a question about the mask of Alphasim correction in voxel-wise functional connectivity. Specifically, to test the link between functional connectivity of the vmPFC and procrastination, I performed a correlation analysis between functional connectivity maps from the vmPFC and scores of procrastination, using result map from one-sample t-test of individuals’ functional connectivity maps of vmPFC as mask. In the following Alphasim correction, Can I use the same result map from one-sample t-tests as mask.
DPABI problems about structural MRI data processing
Dear professor Yan,
I found a problem about structural MRI data processing in DPABI, but im not sure. I found the results of after Image Reslicer(utilities) were binary images...but when i use SPM coreg, it didn't happen.
this is my results after DPABI and SPM8 processing:
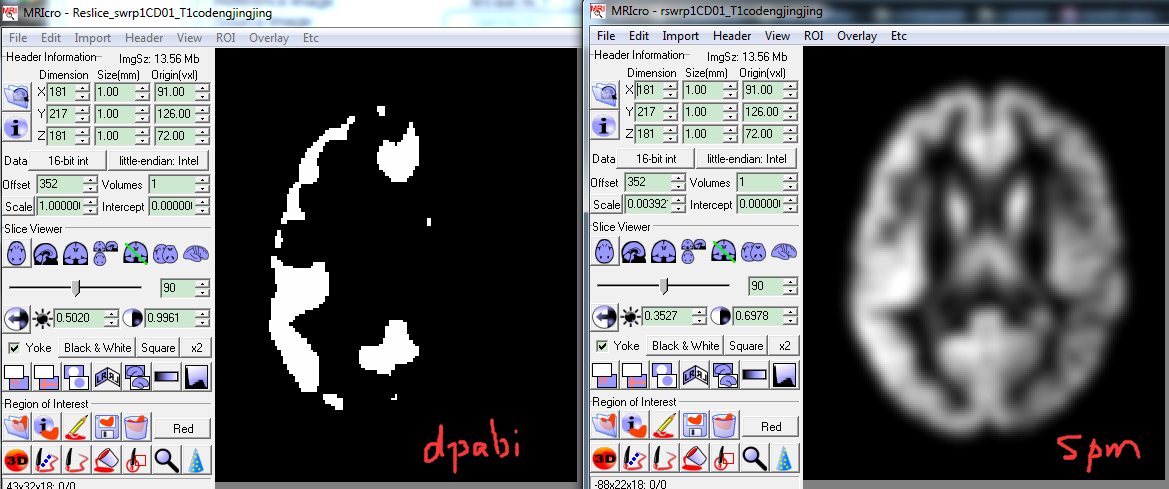
soo, maybe the registeration methods is different.
Thank u for your explanation.
GOOD LUCK
Correct naming of structural folder?
- Read more about Correct naming of structural folder?
- 2 comments
- Log in or register to post comments
Hello,
if using already preprocessed files, I name the folder with the preprocessed functional nifti-files FunImgSARW (smoothed, slice-timing corrected, realigned, normalized). Do I have to rename the structural folder as well (such as e.g. T1ImgRW, because the structural data is not smoothed) or can I keep the name T1Img?
Thanks!
Cheers,
Sascha
NEW GraphVar Release 1.0
- Read more about NEW GraphVar Release 1.0
- Log in or register to post comments
Dear GraphVar community,
we are proud to announce that GraphVar has finally left the beta stage and that the 1.0 release (‘Turbo GLM’) is available as of now on our NITRC page: www.nitrc.org/projects/graphvar
We hope that you will enjoy it as much as we do!
New features include:
Realignment Parameters for already preprocessed files
Dear all,
I am completely new to DPARSFA so this is a very basic question:
