What's the Slice Order of the NKI-RS Lite Releases Data
Hi all,
Could anyone give me some information about the R-fMRI Slice Order of NKI-RS Lite Releases. Thank you in advance!
Best regards,
Huangjing
Hi all,
Could anyone give me some information about the R-fMRI Slice Order of NKI-RS Lite Releases. Thank you in advance!
Best regards,
Huangjing
Given the amount of time and effort for interactive reorienting, here we released the reorient matrices for some of the publicly released data. Please download and unzip the related file, and then put the folder of “DownloadedReorientMats” under your working directory.
1. Autism Brain Imaging Data Exchange (ABIDE) (Di Martino et al., in press) (http://fcon_1000.projects.nitrc.org/indi/abide/). Please download: DownloadedReorientMats_ABIDE.zip.
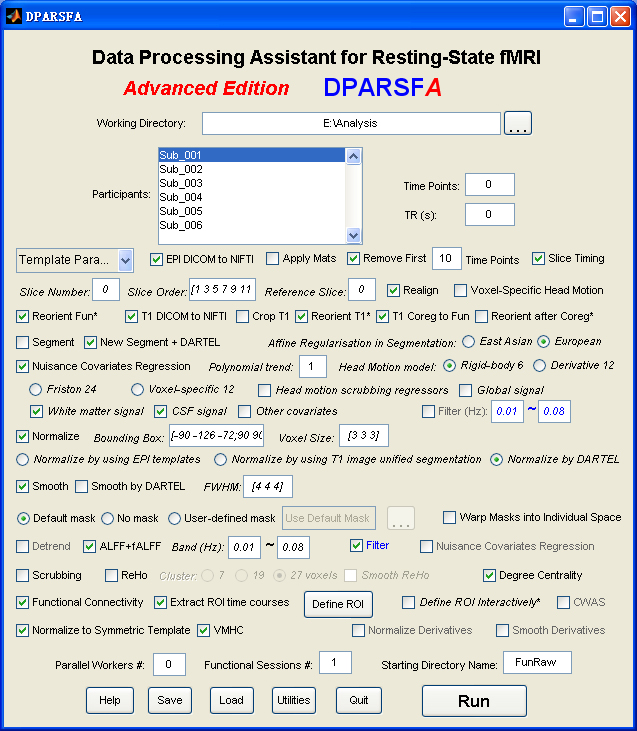 |
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software based on SPM and REST. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, normalize, smooth) data, functional connectivity, ReHo, ALFF/fALFF, degree centrality, voxel-mirrored homotopic connectivity (VMHC) results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract ROI time courses efficiently if you want to perform small-world analysis. DPARSF basic edition is very easy to use while DPARSF advanced edition (alias: DPARSFA) is much more flexible and powerful. DPARSFA can parallel the computation for each subject, and can be used to reorient your images interactively or define regions of interest interactively. You can skip or combine the processing steps in DPARSF advanced edition freely. Please download a MULTIMEDIA COURSEto know more about how to use this software. Add DPARSF's directory to MATLAB's path and enter "DPARSF" or "DPARSFA" in the command window to enjoy DPARSF basic edition or advanced edition. The latest release is DPARSF_V2.3_130615. |
New features of DPARSF_V2.3_130615:
1. Apply downloaded reorient matrices. Given the amount of time and effort for interactive reorienting, the reorient matrices for online data such as 1000 Functional Connectomes Project (FCP) (Biswal et al., 2010) and Autism Brain Imaging Data Exchange (ABIDE) (Di Martino et al., 2013) could be downloaded and applied automatically. Please find here for “DownloadedReorientMats”, and then tick the checkbox "Apply Mats".
2. Save information of TR, Slice Number, Time Points and Voxel Size into TRInfo.tsv (under working directory) file for checking data correctness.
3. The slice order type could be specified for each participant into SliceOrderInfo.tsv (under working directory) file, thus allow different slice timing correction for different participants in a batch mode. Please find instructions for setting SliceOrderInfo.tsv from {DPARSF}/Docs/SliceOrderInfo.tsv_Instruction.txt.
4. The output format of DICOM to NIfTI were changed to 4D .nii images.
5. The midline of VMHC results were set to zero.
Please see the figure below (aal.nii opened by MRIcroN).
The MNI coordinates of the current position is -66 -92 -40, and its value is 0. (-66x-92x-40= 0 on the titile).
The voxel index is 25 34 32. (X 25 Y 34 Z 32).
1. Read the file header:
[Data Header]=y_Read('aal.nii');
2. Convert from voxel index to MNI coordinates:
MNI_Coordinates = Header.mat*[25;34;32;1]
MNI_Coordinates =
-66
-92
-40
1
Dear REST expert, I have a question about the statistic method for the FC analysis. We have a single case vs a group of controls(N = 12). We don't know which test is suitable for the comparison between this single case and a group of controls. Best regards Fede