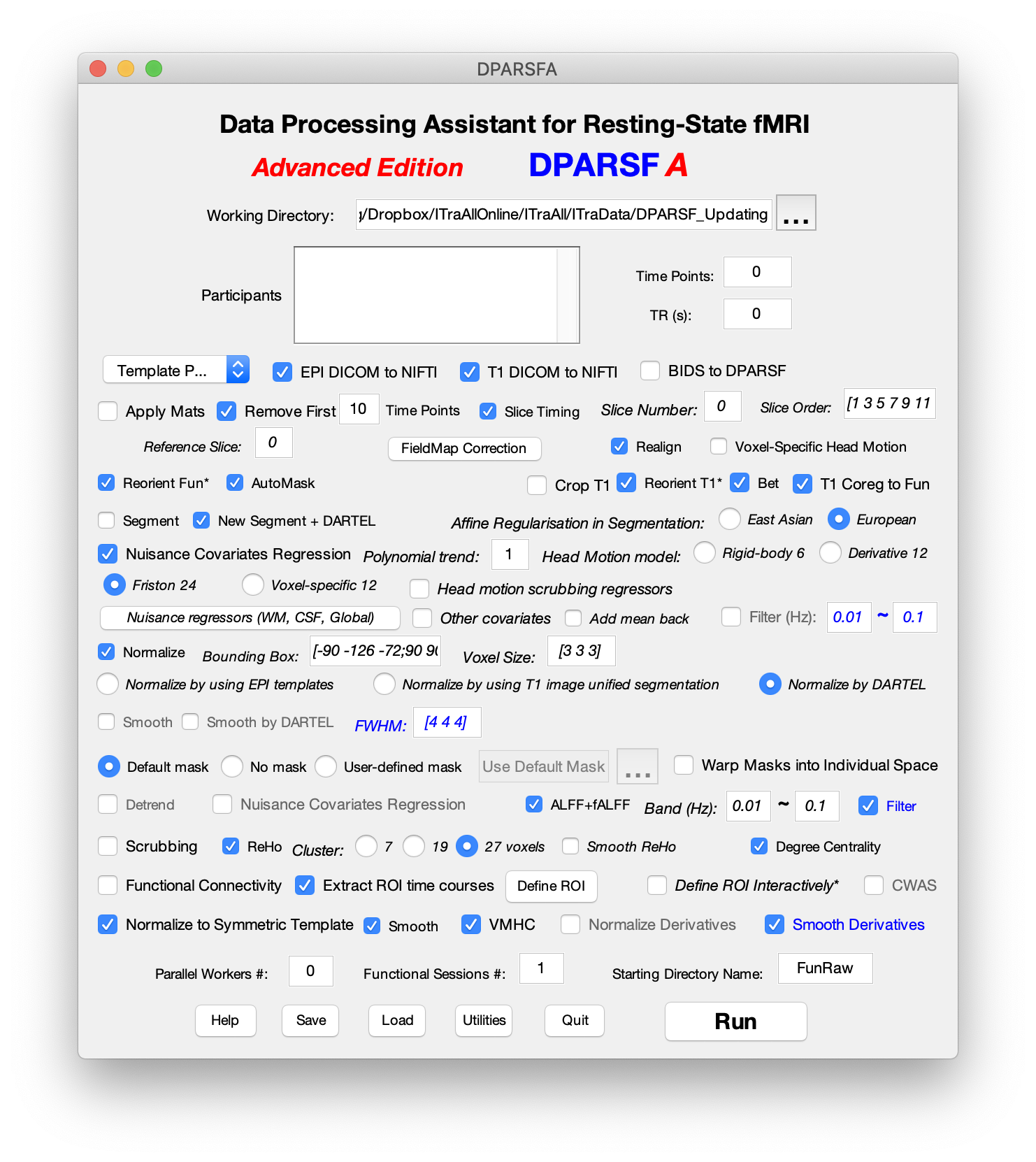
Data Processing Assistant for Resting-State fMRI (DPARSF)
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software within DPABI, which is based on SPM. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, normalize, smooth) data, functional connectivity, ReHo, ALFF/fALFF, degree centrality, voxel-mirrored homotopic connectivity (VMHC) results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract ROI time courses efficiently if you want to perform small-world analysis. DPARSF basic edition is very easy to use while DPARSF advanced edition (alias: DPARSFA) is much more flexible and powerful. DPARSFA can parallel the computation for each subject, and can be used to reorient your images interactively or define regions of interest interactively. You can skip or combine the processing steps in DPARSF advanced edition freely. Please download a MULTIMEDIA COURSE to know more about how to use this software. As a component of DPABI, please add with subfolders for DPABI in MATLAB's path setting and enter "dpabi" in the command window to enjoy this powerful toolbox.
The latest release is DPARSF_V5.3_210101. Please download DPABI to enjoy it!
1. Use 3dAutomask in AFNI to generate automasks if docker is installed.
1. Users can threshold subjects with bad quality after reorienting.
2. Allow skipping subjects in TRInfo.tsv.
Fixed a bug in applying slice timing information from DICOM files to DPARSF settings. This bug only affected DPARSF V5.0 while not setting slice timing information (using the default) in slice timing preprocessing. Automatically applying slice timing information from DICOM files missed the unit change for SPM.
Fixed a bug specific in Windows OS when starting from .nii.gz files: bet fails.
1. Stability Analysis module was added. You can calculate volume-based and surface-based stability from DPABI->Dynamic & Stability Analyses. You can read our recent work for the details of stability measure: Li, L., Lu, B., Yan, C.G. (2019). Stability of dynamic functional architecture differs between brain networks and states. Neuroimage, 116230, doi:10.1016/j.neuroimage.2019.116230.
2. Field map correction was added, both for DPARSF and DPABISurf. If you want to perform field map Correction, you need to arrange each subject's field map DICOM files in one directory, and then put them in "FieldMap" directory under the working directory. i.e.:';...
'{Working Directory}\FieldMap\PhaseDiffRaw\Subject001\xxxxx001.dcm';...
'{Working Directory}\FieldMap\PhaseDiffRaw\Subject001\xxxxx002.dcm';...
'...';...
'{Working Directory}\FieldMap\PhaseDiffRaw\Subject002\xxxxx001.dcm';...
'{Working Directory}\FieldMap\PhaseDiffRaw\Subject002\xxxxx002.dcm';...
'...';...
'{Working Directory}\FieldMap\Magnitude1Raw\Subject001\xxxxx001.dcm';...
'{Working Directory}\FieldMap\Magnitude1Raw\Subject001\xxxxx002.dcm';...
'...';...
'{Working Directory}\FieldMap\Magnitude1Raw\Subject002\xxxxx001.dcm';...
'{Working Directory}\FieldMap\Magnitude1Raw\Subject002\xxxxx002.dcm';...
'...';...
'...';...
Then you can click the button of “FieldMap” button to set field map correction parameters. In most cases, you can use the default “0” value to let the program read the parameters (e.g., echo times) from the DICOM files.
3. Check data organization function added. For the new users of DPARSF and DPABISurf, most of the errors were caused by data organization! Please use DPABI->Utilities->Check Data Organization to check your data organization before running DPARSF or DPABISurf. This program will lead you to organize your data correctly with prompting messages!
4. Slice timing information read from DICOM files. If you are starting with DICOM files, you no longer need to set the slice timing correction parameters. Just leave it as default (slice number: 0), then DPARSF or DPABISurf will read the parameters from DICOM files. This new feature thanks to Dr. Chris Rorden's new dcm2niiX program (version v1.0.20190902).
5. DPARSF V5.0 now is compatible with BIDS format. You can start with BIDS format data by checking checkbox “BIDS to DPARSF” and setting “Starting Directory Name” to “BIDS”.
1. For Linux or Mac OS, if FSL is not installed, then DPARSF will call FSL's bet in dpabi docker.
Tips for Linux or Mac O: please start matlab from terminal in order to reach docker in DPABI (e.g., Linux: matlab; Mac: open /Applications/MATLAB_R2018a.app/).
New features of DPARSF_V4.4_180801 (download at http://rfmri.org/dpabi):
1. Added a prompt of "Congratulations, the running of DPARSFA is done!!! :)" when DPARSF finishes its processing.
2. Added a new atlas (Schaefer2018_400Parcels_7Networks_order_FSLMNI152_1mm.nii) to the V4 parameters. Please see more details at Schaefer, A., Kong, R., Gordon, E.M., Laumann, T.O., Zuo, X.N., Holmes, A.J., Eickhoff, S.B., Yeo, B.T.T., 2017. Local-Global Parcellation of the Human Cerebral Cortex from Intrinsic Functional Connectivity MRI. Cereb Cortex, 1-20.
3. The dcm2nii has been updated to the latest version in courtesy of Dr. Chris Rorden. See: Li, X., Morgan, P.S., Ashburner, J., Smith, J., Rorden, C., 2016. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods 264, 47-56.
4. As there were some parallel computing issues in calling outside command, the callings were no longer using parallel computing (i.e., downgrade from parfor to for). These includes the callings of dcm2nii and bet.
5. Flexibility for concordance was added to the module of Temporal Dynamic Analysis (DPABI_TDA). Users can freely calculate the concordance of any combinations of ALFF, fALFF, ReHo, Degree Centrality, Global Signal Correlation and VMHC.
6. Fixed some compatibility bugs with higher versions of MATLAB. For example, Time Course error in DPABI_VIEW; uimenu parent problem when calling monkey/rat module; errors regard generating pictures for checking normalization in DPARSFA.
7. Tips for calling "bet": You should start matlab from terminal (e.g., Linux: matlab; Mac: open /Applications/MATLAB_R2018a.app/). If you installed FSL5.0, you may also need to run this: source /usr/share/fsl/5.0/etc/fslconf/fsl.sh. In addition, in some Linux versions, you may need to start matlab in this way: LD_PRELOAD="/usr/lib/x86_64-linux-gnu/libstdc++.so.6" matlab.
New features of DPARSF_V4.3_170105:
1. DPARSFA V4 Parameters (Default Parameters, also for The R-fMRI Maps Project). For ROI signals extraction, the Power 264 ROIs were added as the 1570~1833 ROIs. (Power_Neuron_264ROIs_Radius5_Mask.nii was added to {DPABI}/Templates/)
2. DPARSF Advanced Edition: Re-run with global signal regression (DPARSFA_RerunWithGSR). Fixed a bug when “Remove first X time points” was defined, the number of time points will be adjusted accordingly.
3. DPARSF. Add a “Clear All” button in the ROI List GUI for defining ROIs.
New features DPARSF_V4.2_161201
1. To let the users be more aware what kind of templates they are using, SPM Templates were included under {DPABI}/Templates/ now. If you want to USE YOUR OWN TEMPLATES, please replace the corresponding ones under this directory instead of replacing those under SPM. For example: if you are using normalize by New Segment + DARTEL, please replace {DPABI}/Templates/SPMTemplates/tpm/TPM.nii; If you are using normalize by using EPI template, please replace {DPABI}/Templates/SPMTemplates/toolbox/OldNorm/EPI.nii; If you are using normalize by using T1 image unified segmentation, please replace {DPABI}/Templates/SPMTemplates/toolbox/OldSeg/grey.nii, white.nii, and csf.nii.
2. DPARSF Windows version. Previously need to run as administrator to get results both with and without global signal regression (GSR). Now such limitation is removed (change mklink to copyfile).
3. DPARSF Advanced Edition Preprocessing for Task fMRI data: For nuisance regression, the option of “Add mean back” is now default. The mean will be added back to the residual after nuisance regression. This is useful for circumstances of ICA or task-based analysis.
4. DPARSFA V4 Parameters (Default Parameters, also for The R-fMRI Maps Project). For ROI signals extraction, the global signal (BrainMask_05_91x109x91.img) was added as the 1569th ROI.
New features of DPARSF V4.1_160415:
1. Fixed a bug in DPARSF Basic Edition. The bug is that the white matter signal is always removed in nuisance regression (only exist in the Basic Edition). Thanks to the report of Liviu Badea.
2. DPARSF Advanced Edition: Add an option of “Add mean back” for nuisance regression. The mean will be added back to the residual after nuisance regression. This is useful for circumstances of ICA or task-based analysis.
3. DPARSF Advanced Edition: Re-run with global signal regression (DPARSFA_RerunWithGSR). Fixed a bug when “Remove first X time points” was defined, the number of time points will be adjusted accordingly now. Thanks to the report of Hua-Sheng Liu.
4. DPARSF Advanced Edition: Add a slice timing batch mode for MultiBand data. Users could specify a text timing file for a given participant in SliceOrderInfo.tsv. Please see http://rfmri.org/SliceTiming for more details.
New features of DPARSF_V4.0_151201 (together with DPABI_V2.0_151201) :
1. Compatible with MATLAB 2014b and later versions.
2. Process the data both with and without global signal regression (GSR). Check “Nuisance regressors setting” -> “Both with & without GSR”. Alternatively, you can call DPARSFA_RerunWithGSR.m. E.g., DPARSFA_RerunWithGSR(DPARSFACfg.mat); where DPARSFACfg.mat stores the previous parameters without GSR.
3. The processing steps are affixed to Results directories. The R-fMRI calculation parameters are also written to the header of the result files.
4. V4 template is added. No smoothing before R-fMRI measure calculation (except for VMHC). This is used for comparing across studies and accumulate processed data.
5. DPABI Statistical Analysis. Add multiple comparison test after ANOVA, e.g., 'tukey-kramer' or 'hsd', 'lsd', 'dunn-sidak', 'bonferroni’ or ‘scheffe' procedures.
6. DPABI_VIEW: compatible with BrainNet Viewer 1.5.
7. Fixed a "File too small" bug when .hdr/.img files are used.
8. Fixed a bug in y_Standardize.m: error when multiple files are defined.
9. Fixed a bug in DPABI Image Calculator: error in standard deviation calculation along the 4th dimension.
10. Results Organizer module: with this module, the users could organize the intermediate files for future processing with DPABI. In addition, the results could be organized for future use, and to be accumulated for the future R-fMRI maps project.
New features of DPARSF_V3.2_150710:
1. SPM12 Compatible.
2. DPARSF for Rat data released.
The Rat module is based on a Rat T2 template generated by Dr. Adam J. Schwarz et al. Please cite this paper when appropriate: Schwarz, A.J., Danckaert, A., Reese, T., Gozzi, A., Paxinos, G., Watson, C., Merlo-Pich, E.V., Bifone, A., 2006. A stereotaxic MRI template set for the rat brain with tissue class distribution maps and co-registered anatomical atlas: application to pharmacological MRI. Neuroimage 32, 538-550. (A T1 template was included as well. It's generated by normalizing 50 rats (two scans at PND45 or PND60) to that T2 template and then averaging (by Dr. Chao-Gan Yan)).
A video for rat data processing is available at http://d.rnet.co/DPABI_RatDataProcessing_20150520.mp4.
3. Fixed a bug in generating Voxel Specific Head Motion: missing gmdmp.
New features in DPARSF_V3.1_141101.
1. DPARSF V3.1 Basic Edition: Fixed a bug of missing DPARSF_run.
2. DPARSF V3.1: Fixed a bug that can not find ROI templates.
1. New features in DPARSF 3.0 Advanced Edition.
1.1. Quality control. Integrated GUI for QCing the functional and structural images, users can give ratings and comments during the step of interactive reorientation.
1.2. Automask generation. For checking EPI coverage and generating group mask, the automasks (as in AFNI) will be generated based on EPI images.
1.3. Brain extraction (Skullstrip). This step can improve the coregistration between functional and structural images. Most registration issues of previous DPARSF versions can be solved by including this step. For Linux and Mac users: Need to install FSL. For Windows users: Thanks to Chris Rorden's compiled version of bet in MRIcroN, the modified version can work on NIfTI images directly.
1.4. Nuisance Regression. 1) Masks can be generated based on segmentation or SPM apriori masks; 2) Methods can be mean or CompCor [Note: for CompCor, detrend (demean) and variance normalization will be applied before PCA, according to (Behzadi et al., 2007)]; 3) Global Signal can be extracted based on Automasks.
2. New features in DPARSF 3.0 Basic Edition.
2.1. DPARSF Basic Edition now is using the engine of DPARSF Advanced Edition.
2.2. Nuisance Regression (in MNI space) is placed before filtering, according to (Hallquist et al., 2013).
3. New features in DPARSF for Monkey data.
3.1. The monkey module is based on Rhesus Macaque Atlases for functional and structural imaging studies generated by Wisconsin ADRC Imaging Core. Please cite their papers when appropriate: (McLaren et al., 2010; McLaren et al., 2009).
3.2. Of note, the origin of monkey atlas is different from human MNI atlas. Please make sure the correct origins are set at the steps of "reorienting Fun*" and "reorienting T1*".
Licence: GNU General Public Licence (GPL)


there is an error in my preprocessing procedure.
Hi,
Dr Yan,
there is an error in my preprocessing procedure as below, My spm is version 8 and the matlab version is R2010b, is there any problem about this ? Thank you!
Hi,
Hi,
Please re-add with subfolders for SPM8 path.
Best,
Chao-Gan
Error in ==> file_array.subsref>subfun at 80
RE: [RFMRI] Data Processing
Sent: Tuesday, December 01, 2015 8:59 PM
To: rfmri.org@rnet.co
Subject: Re: [RFMRI] Data Processing Assistant for Resting-State fMRI (DPARSF) V4.0
Hi,
Hi,
One .nii is OK, as it contains 240 volumes.
The error you encountered is caused by the conflict between REST and SPM8. REST contains some spm5 files, thus please DO NOT add with subfolders for REST. Or remove REST path to have a try.
Best,
Chao-Gan
RE: [RFMRI] Data Processing
Fwd: [RFMRI] Data Processing
"Too many .nii.gz files in each subject's directory" error
Hello,
I am new to DPARSFA and I hope you can help me. I got the following error after specifying my working directory and starting directory (FunImg): "Too many .nii.gz files in each subject's directory, should only keep one 4D .nii.gz file". However, it is only a test run with one subject, so I have one .nii file for this subject in FunImg folder and one .nii.gz file for this subject in the T1Img folder (so one file in each folder in total). What can cause the problem?
Thanks a lot!
Lisa
Can you help me?
Hi,
Dr Yan,
I have an error in my preprocessing procedure for resting state as below, I use spm8 、 matlab 2007a and DPARSF_V2.3。Is there any problem about this ? Thank you!
Running 'New Segment'
Failed 'New Segment'
Error using ==> zeros
Out of memory. Type HELP MEMORY for your options.
In file "F:\MATLAB\R2007a\work\spm8\toolbox\Seg\spm_preproc_write8.m" (v2531), function "rgrid" at line 435.
In file "F:\MATLAB\R2007a\work\spm8\toolbox\Seg\spm_preproc_write8.m" (v2531), function "spm_preproc_write8" at line 241.
In file "F:\MATLAB\R2007a\work\spm8\toolbox\Seg\spm_preproc_run.m" (v2281), function "run_job" at line 112.
In file "F:\MATLAB\R2007a\work\spm8\toolbox\Seg\spm_preproc_run.m" (v2281), function "spm_preproc_run" at line 27.
The following modules did not run:
Failed: New Segment
??? Error using ==> cfg_util.m>local_runcj at 251
Job execution failed. The full log of this run can be found in MATLAB command window, starting with the lines (look for the line showing the exact #job as displayed in this error message)
------------------
Running job #1
------------------
Error in ==> DPARSFA>pushbuttonRun_Callback at 1601
[Error]=DPARSFA_run(handles.Cfg);
Error in ==> gui_mainfcn at 95
feval(varargin{:});
Error in ==> DPARSFA at 33
gui_mainfcn(gui_State, varargin{:});
??? Error using ==> DPARSFA('pushbuttonRun_Callback',gcbo,[],guidata(gcbo))
Error using ==> cfg_util.m>local_runcj at 251
Job execution failed. The full log of this run can be found in MATLAB command window, starting with the lines (look for the line showing the exact #job as displayed in this error message)
------------------
Running job #1
------------------
??? Error while evaluating uicontrol Callback
>>
Hi,
Hi,
This is an out of memory error. Please refer to http://rfmri.org/FAQ #2.
Best,
Chao-Gan
dparsf报错
严老师,您好!
我那个dparsf一运行就报错
并且,MATLAB还会有如下提示
想问下您这是怎么回事?要怎么解决呢?
还有,数据的TR不一样,还能批量处理吗?
谢谢
1. 试着安装一下最新的DPABI/DPARSF。
1. 试着安装一下最新的DPABI/DPARSF。
2. 可以批量处理。请见:http://wiki.rfmri.org/content/repetition-time-tr。
严老师,
严老师,
谢谢您之前看回答。
之前的问题已经解决了,但是又出现了新的问题。
dparsf运行到后面就会报错,错误如下
Seems you don't have
Seems you don't have permission on your windows computer to execute "mklink"?
谢谢
谢谢
严老师,我用网站提供的数据就能正常运行
严老师,我用网站提供的数据就能正常运行,但换成我自己的数据时,做完NewSegment之后就报错了……
error in progress
Hi,
This is the error show in my preprocessing procedure as below, My dpabi version is 4.0,spm is version 8 and the matlab is R2013b, is there any problem about this ?
Error using nifti/subsref>rec (line 104)
Thank you!
Is SPM running properly?
Is SPM running properly?
thanks chaogan,
thanks chaogan,
The problem has been sloved. It seems you need to open the SPMwhen running DPARSF.
Best
严老师,您好!
严老师,您好!
我想问下,我在workingdirectory里放入了downloadedreorientmats,选了applymats,但为什么reorient那一步还是要手动调整呢?
已经知道了……
已经知道了……
VMHC results
Hello,
I was using DPARFS to calculate VMHC for resting state fMRI.
I got the results but they look like a mask!
What could i have done wrong? What should i do to fix this?
thanks in advance,
Walid
This is weird. Please check
This is weird. Please check your data input for VMHC calculation.
Hi,
Hi,
Yes, i know it's weird which is why i am asking!
Could you please be more precise? Is there somewhere i should be looking that might have given this output?
I checked my input and i think it is Ok..
Any helpful advice on the matter is highly appreicated!
What's your input for VMHC
What's your input for VMHC calculation. E.g., the starting directory name.
Thank you for your reply. I
Thank you for your reply. I understand.
I will share with you the file structure.
dpabi
——————————
Inside dpabi folder
ch2.nii.gz
dirlist.txt
DPARSFA_setting.mat
F.nii
FunRaw
FunRawR
FunRawRC
FunRawRCovs
FunRawRCW
FunRawRCWS
FunRawRCWSF
FunRawRCWSFsym
Masks
PicturesForChkNormalization
RealignParameter
Results
SymmetricGroupT1MeanTemplate
T1Img
T1ImgBet
T1ImgCoreg
T1ImgSegment
T2.nii
————————
Inside the Results folder
ALFF_FunRawRCWS
DegreeCentrality_FunRawRCWSF
fALFF_FunRawRCWS
results
VMHC_FunRawRCWSFsym
——————————
Inside the results folder
DCDegreeCentrality_FunRawRCWSF.nii
VMHCVMHC_FunRawRCWSFsym.nii
zDCDegreeCentrality_FunRawRCWSF.nii
zVMHCVMHC_FunRawRCWSFsym.nii
If you would like to see the setting file, please let me know where to send it because i couldnt attach it here.
Thank you again,
Walid
Hi
Hi
Inside VMHC_FunRawRCWSFsym
Should be something like this:
VMHCMap_Sub_001.nii
Hello,
Hello,
Inside VMHC_FunRawRCWSFsym i have files (See below)
Inside each file i have
FunRawRCWSFsym/b102/sym_Filtered_4DVolume.mat
The fiels seems right. Could
The fiels seems right. Could you have a test run on the sample data at http://rfmri.org/DemoData?
The fiels seems right. Could
The fiels seems right. Could you have a test run on the sample data at http://rfmri.org/DemoData?
SPM12: spm_coreg: Warning: Too many optimisation iterations
Hi,
I am using DPARSFA on a Mac with OS Sierra. Unfortunately, I cannot get my segmentation to work (NewSegment+DARTEL) and I am already getting a warning during coregistration:
Warning: Too many optimisation iterations
> In spm_powell at 46
I guess there might be some
I guess there might be some problem with your data. Try the demo data first.
严老师:
请问该如何解决呢?之前用SPM预处理数据后得到的图像的dimensions的确是61*73*61。
谢谢
你用的什么参数?模板是3x3x3的吗?
你用的什么参数?模板是3x3x3的吗?
老师,您好!这是我设置的参数:
你用新版本试试?
你用新版本试试?
Warnings of DPARSF4.3
Hello, Dr. Yan,
When I used the DPARSF for preprocessing, in the DARTEL, there were many warnings:
You can ignore them.
You can ignore them.
looking for wAutoMask files - FIXED
edit 2017-06-27 T 1457 PDT
It looks like the problem was at some point I unchecked Normalize by mistake, which I realized after loading the autosave*.mat file and seeing that the configuration was slightly different than the screenshot below. Right now I am re-running this, hopefully the problem is fixed.
Thanks,
Felix
Hello,
Can someone please help me understand where I can find the wAutoMask files? I ran only preprocessing steps so far (please see screenshot), and wanted to create a group mask before postprocessing. Preprocessing appears to have completed with no problem, resulting in swCovRegressed_4DVolume.nii files within FunImgARCFS\SubjID folders. Do the settings I used result in no wAutoMask files being created, and if so, could you recommend an alternative file type or approach to creating a group mask?
Thanks in advance,
Felix
error after realigment step
Hello,
I have used your software to preprocess resting-state data over the years, and your automated preprocessing pipeline DPARSF works very well. I am trying to replicate a finding from my disseration analyses in a new dataset. I have 287 subjects, but did a test on the first three subjects to make sure the automated preprocessing ran smoothly with the latest version of DPARSF (running off DPABI) available on your website. For the test, I selected simple pre-processing: remove first 10 timepoints, slice timing, realignment/motion correction, normalization to EPI template and smoothing at 8 FWHM. DPARSF successfully performed the slice timing and realignment, but then I got this error (see red text below), and it did not complete preprocessing. Do you know what this issue might be? Thanks!
~Stephanie Hare
------------------------------------------------------------------------
Running job #1
------------------------------------------------------------------------
Running 'Slice Timing'
SPM8: spm_slice_timing (v4310) 16:08:36 - 11/10/2018
========================================================================
Your TR is 2.0
Done 'Slice Timing'
Done
Slice Timing Setup:S0229 OK
------------------------------------------------------------------------
Running job #1
------------------------------------------------------------------------
Running 'Slice Timing'
SPM8: spm_slice_timing (v4310) 16:18:26 - 11/10/2018
========================================================================
Your TR is 2.0
Done 'Slice Timing'
Done
Slice Timing Setup:S0230 OK
------------------------------------------------------------------------
Running job #1
------------------------------------------------------------------------
Running 'Slice Timing'
SPM8: spm_slice_timing (v4310) 16:30:13 - 11/10/2018
========================================================================
Your TR is 2.0
Done 'Slice Timing'
Done
Moving Slice Timing Corrected Files:S0230 OKMoving Slice Timing Corrected Files:S0229 OKMoving Slice Timing Corrected Files:S0226 OK
Realign Setup:S0226 OK
------------------------------------------------------------------------
Running job #1
------------------------------------------------------------------------
Running 'Realign: Estimate & Reslice'
Done 'Realign: Estimate & Reslice'
Done
Realign Setup:S0229 OK
------------------------------------------------------------------------
Running job #1
------------------------------------------------------------------------
Running 'Realign: Estimate & Reslice'
Done 'Realign: Estimate & Reslice'
Done
Realign Setup:S0230 OK
------------------------------------------------------------------------
Running job #1
------------------------------------------------------------------------
Running 'Realign: Estimate & Reslice'
Done 'Realign: Estimate & Reslice'
Done
Moving Head Motion Corrected Files:S0230 OKMoving Head Motion Corrected Files:S0229 OKMoving Head Motion Corrected Files:S0226 OK
Reorienting Functional Images Interactively for S0226:
Reference to non-existent field 'ReorientMat'.
Error in DPARSFA_run (line 1196)
mat=y_spm_image_Parameters.ReorientMat;
Error in DPARSF_run (line 113)
[Error] = DPARSFA_run(Cfg);
Error in DPARSF>pushbuttonRun_Callback (line 1154)
[Error]=DPARSF_run(handles.Cfg);
Error in gui_mainfcn (line 96)
feval(varargin{:});
Error in DPARSF (line 52)
gui_mainfcn(gui_State, varargin{:});
Error using drawnow
Error while evaluating uicontrol Callback
Maybe you need to check the
Maybe you need to check the "reorient fun" and "reorient t1" checkboxes and reorient your images.
体素级脑功能网络相关矩阵
严老师,
您好!我想做的是基于DPARSF软件,将预处理后的脑功能网络图像进行基于体素的相关分析,对于每一被试,计算全脑任意一对体素时间序列间的Pearson相关系数,生成一个高维(如70831*70831)的相关矩阵。目前,我可以提取到如AAL的90*90的相关矩阵,Power_Neuron_264ROIs的264*264的相关矩阵,但提取不到高维的关于体素级的相关矩阵。这个高维的体素级脑网络相关矩阵该如何提取到?
You can use y_ReadAll.m read
You can use y_ReadAll.m read the data and calculate by yourself. Read more here: http://lab.rfmri.org/Course/V2.3CH/12_BrainImagingProgramming.mp4
Hi
Hi
I faced an error processing my data. while it was converting t1 dicom to nifti, I got this error
I am using MATLAB R2018b and SPM12 . my data is from ADNI database.
would you please help me?
Please check very carefully
Please check very carefully your FunImg directory about 1) whether there is empty folder 2) whether there are folders that contain different number of dicom files
About Auto MASK error
Checking off Automask button
Checking off Automask button should be OK. What's the error message then?
Checking off Automask button
Checking off Automask button should be OK. What's the error message then?
error "insufficient image overlap"
Hi Dr. Yan,
I am running the "Normalize to Symmetric Template" before the VMHC analysis and meet an error as followed for some of my subjects :