I want to obtain network measurements that illustrate the group comparisons I’m performing on my data. The data itself consists of coherence matrixes previously calculated by Matlab routines external to GraphVar. So, in order to work with them, I’m selecting the “Raw Matrix” checkbox in the “Raw Matrix (link wise)” panel.
For this exposition, let’s focus solely on the computation of the “Binary Degree” of each node. I imposed the relative network thresholds of 0.1, 0.2, 0.3, 0.4 and 0.5 in the “Network Construction” menu.
My problem occurs when I select the “Normalize graph metric with random networks”.
For faster computation, I’m restringing the trial to 22 brain areas, shortening the repetitions (“Test against random groups” = 2) and the number of the null model networks created (5 network created with the randomizer_bin_und function with only 1 iteration).
If the program does run, in the proposed configuration (see the interface matching the description portrayed below), I get the following results:


Results that don’t seem to produce valid information. The values of “Degree” for a given area and respective associated p-value just show two possibilities: either 0 and NaN (green), or NaN and NaN (blue).
Now, for, instance, just by slightly increasing the “Number of Repetitions” to 10 in the “Test against random groups option” (“Statistics” panel), the software crashes when it appears to be correlating the data:
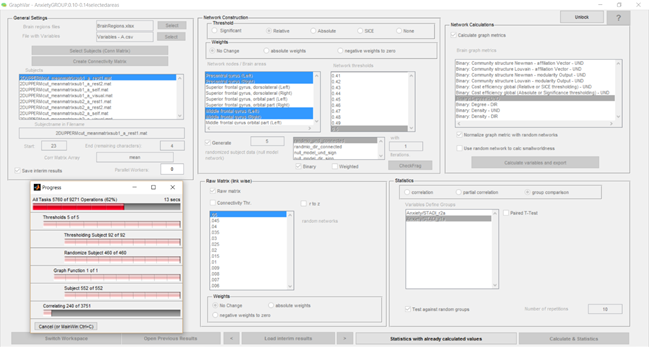
Presenting the following error:
Operands to the || and && operators must be convertible to logical scalar values.
Error in idummy (line 22)
if (g > 1 || method==3)
Error in anovan>makedummyvars (line 1242)
vdum{j} = idummy(allgrps(:,j), 3);
Error in anovan (line 185)
varinfo = makedummyvars(continuous,allgrps,vnested,varinfo);
Error in executeStats>StatsFunction (line 430)
[Result.anovaP,tab] = anovan(brainData,{variable},'display','off');
Error in executeStats>nDimStats (line 316)
Result = StatsFunction(fid,subjectList,brainData,variable,StatsType,partialVar,isRand,0);
Error in executeStats>nDimStats (line 324)
eval(['[res,Result(' dimStr 'i)] =
nDimStats(fid,subjectList,actTask,brainDataNew,variable,StatsType,partialVar,isRand,loopCountOut);']);
Error in executeStats (line 97)
[res, Result] =
nDimStats(fid,subjectList,actTask,GraphVar.Result,VarData,statsType,[],i_run >
1);
Error in GraphVar_corr (line 89)
[Files,N] =
executeStats(allTasks,dialogData.statsType,dialogData.functionList,dialogData.thresholds,dialogData.VarList,dialogData.CovarList,nRandom,dialogData.nShuffel,0,dialogData.ConnectivityT
Error in GraphVar>Btn_calcCorr_Callback (line 75)
GraphVar_corr(handles,allTask)
Error in gui_mainfcn (line 96)
feval(varargin{:});
Error in GraphVar (line 40)
gui_mainfcn(gui_State, varargin{:});
Error in @(hObject,eventdata)GraphVar('Btn_calcCorr_Callback',hObject,eventdata,guidata(hObject))
Error while evaluating uicontrol Callback
On the other hand, if I don’t click on the checkbox “Normalize graph metric with random networks”, the program runs as I expected it to run. Below are the prints of the interface and the result box obtained:
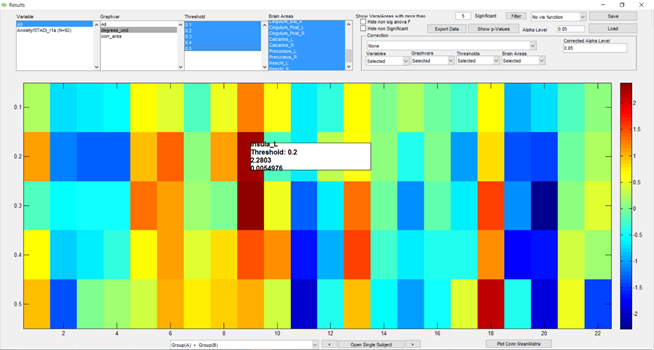
So, in resume, I’m doubting my usage of GraphVar on the following parameters:
Am I using the software properly in order to achieve the “Degree” measurement with the significance values calculated with the null-model networks I created? Or is the “Normalize graph metric with random networks” option not meant to be used this way (when performing group comparisons)?
Also, are the generated null-model networks being used in any way for the calculation of the significance values of the network metrics selected if I opt out of clicking on the “Normalize graph metric with random networks” checkbox?

"Normalize graph metric with random networks” option
Hi,
the problem occurs as your networks seem to be fragmented, which results in some nodes of degree 0. If you subsequently normalize with the node degrees of the random networks (original degree/mean (degree random networks)) you will receive a NaN as division by 0 is not possible. By the way: the randomization functions will keep the basic properties of the random network the same as the original networks... therefore a node degree from these random networks will be the same as the degree of the same node in the original non-randomized data (i.e., the number of connections stays the same but it connected nodes will vary). Thus, if your network is not fragmented normalization by node degree from random networks will necessarily result in 1 for all nodes (please have a look at the BCT functions where this is explained).
The other error you get when you set the "test against random groups" parameter to 10 seems to result from the statistics toolbox itself (i.e., from the anova function). Here, I cannot say what goes wrong at the moment without insight into your data structure.
Hope this helps,
Johann