The toolbox for Data Processing & Analysis for Brain Imaging on Surface (DPABISurf) (Yan et al., 2021) is based on fMRIPrep (Esteban et al., 2019), FreeSurfer (Dale et al., 1999), ANTs (Avants et al., 2008), FSL (Jenkinson et al., 2002), AFNI (Cox, 1996), SPM (Ashburner, 2012), dcm2niix (Li et al., 2016), PALM (Winkler et al., 2016), GNU Parallel (Tange, 2011), MATLAB (The MathWorks Inc., Natick, MA, US), Docker (https://docker.com) and DPABI (Yan et al., 2016). DPABISurf provides user-friendly graphical user interface (GUI) for pipeline surface-based preprocessing, statistical analyses and results viewing, while requires no programming/scripting skills from the users.
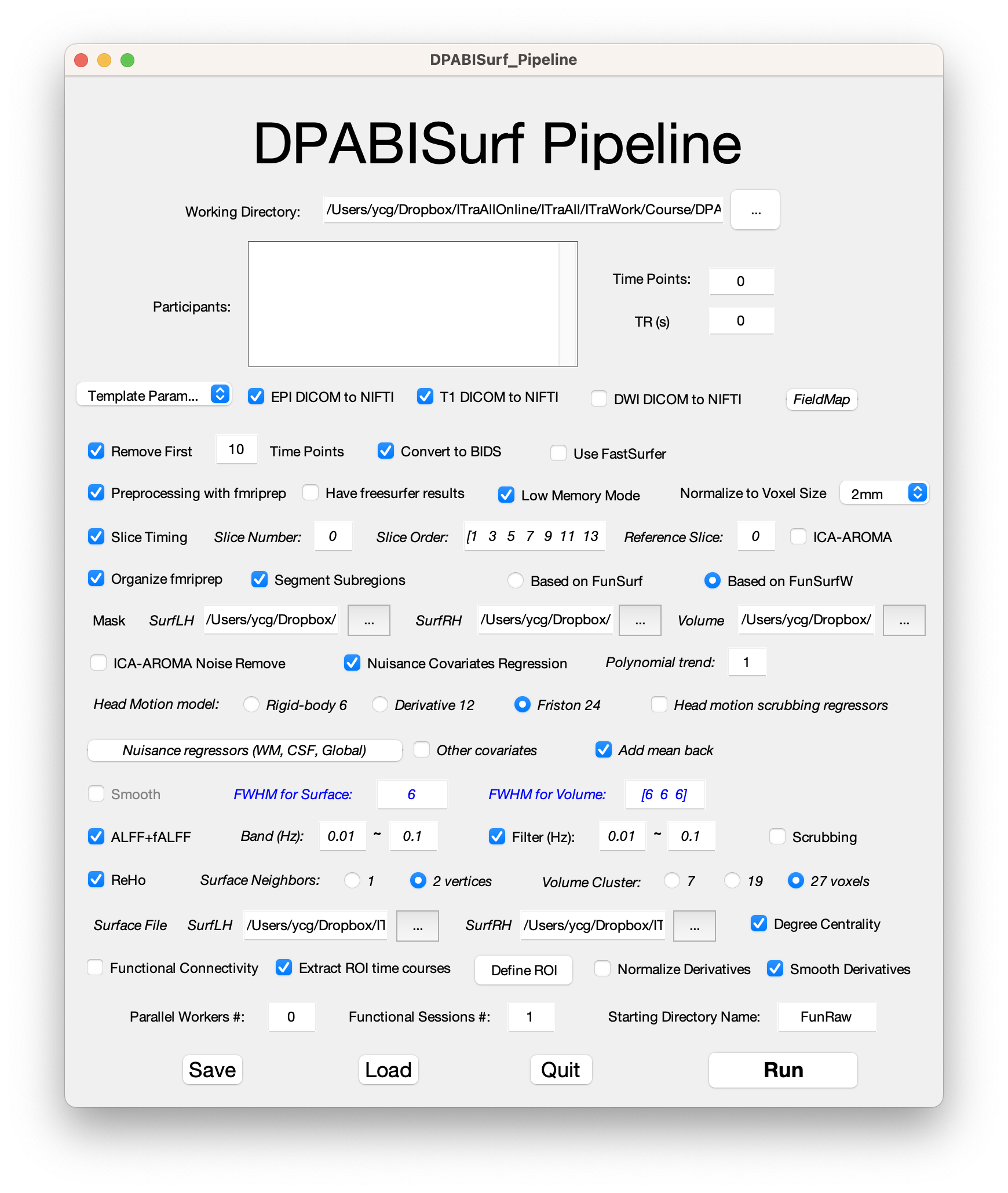
The DPABISurf pipeline first converts the user specified data into BIDS format (Gorgolewski et al., 2016), and then calls fMRIPprep docker to preprocess the structural and functional MRI data, which integrates FreeSurfer, ANTs, FSL and AFNI. With fMRIPprep, the data is processed into FreeSurfer fsaverage5 surface space and MNI volume space. DPABISurf further performs nuisance covariates regression (including ICA-AROMA) on the surface-based data (volume-based data is processed as well), and then calculate the commonly used R-fMRI metrics: amplitude of low frequency fluctuation (ALFF) (Zang et al., 2007), fractional ALFF (Zou et al., 2008), regional homogeneity (Zang et al., 2004), degree centrality (Zuo and Xing, 2014), and seed-based functional connectivity. DPABISurf also performs surface-based smoothing by calling FreeSurfer’s mri_surf2surf command. These processed metrics then enters surfaced-based statistical analyses within DPABISurf, which could perform surfaced-based permutation test with TFCE by integrating PALM. Finally, the corrected results could be viewed by the convenient surface viewer DPABISurf_VIEW, which is derived from spm_mesh_render.m.
DPABISurf is designed to make surface-based data analysis require minimum manual operations and almost no programming/scripting experience. We anticipate this open-source toolbox will assist novices and expert users alike and continue to support advancing R-fMRI methodology and its application to clinical translational studies.
DPABISurf is open-source and distributed under GNU/GPL, available with DPABI at http://www.rfmri.org/dpabi. It supports Windows 10 Pro, MacOS and Linux operating systems. You can run it with or without MATLAB.
1. With MATLAB.
1.1. Please go to http://www.rfmri.org/dpabi to download DPABI.
1.2. Add with subfolders for DPABI in MATLAB's path setting.
1.3. Input 'dpabi' and then follow the instructions of the "Install" Button on DPABISurf.
2. Without MATLAB.
2.1. Install Docker.
2.2. Terminal: docker pull cgyan/dpabi
2.3. Terminal: docker run -d --rm -v /My/FreeSurferLicense/Path/license.txt:/opt/freesurfer/license.txt -v /My/Data/Path:/data -p 5925:5925 cgyan/dpabi x11vnc -forever -shared -usepw -create -rfbport 5925
/My/FreeSurferLicense/Path/license.txt: Where you stored the FreeSurferLicense got from https://surfer.nmr.mgh.harvard.edu/registration.html.
/My/Data/Path: This is where you stored your data. In Docker, the path is /data.
2.4. Open VNC Viewer, connect to localhost:5925, the password is 'dpabi'.
2.5. In the terminal within the VNC Viewer, input "bash", and then input:
/opt/DPABI/DPABI_StandAlone/run_DPABI_StandAlone.sh ${MCRPath}
Now please enjoy the StandAlone version of DPABISurf with GUI!
References:
· Ashburner, J. (2012). SPM: a history. Neuroimage, 62(2), 791-800, doi:10.1016/j.neuroimage.2011.10.025.
· Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal, 12(1), 26-41, doi:10.1016/j.media.2007.06.004.
· Cox, R.W. (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res, 29(3), 162-173.
· Dale, A.M., Fischl, B., Sereno, M.I. (1999). Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage, 9(2), 179-194, doi:10.1006/nimg.1998.0395.
· Esteban, O., Markiewicz, C.J., Blair, R.W., Moodie, C.A., Isik, A.I., Erramuzpe, A., Kent, J.D., Goncalves, M., DuPre, E., Snyder, M., Oya, H., Ghosh, S.S., Wright, J., Durnez, J., Poldrack, R.A., Gorgolewski, K.J. (2019). fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods, 16, 111-116, doi:10.1038/s41592-018-0235-4.
· Gorgolewski, K.J., Auer, T., Calhoun, V.D., Craddock, R.C., Das, S., Duff, E.P., Flandin, G., Ghosh, S.S., Glatard, T., Halchenko, Y.O., Handwerker, D.A., Hanke, M., Keator, D., Li, X., Michael, Z., Maumet, C., Nichols, B.N., Nichols, T.E., Pellman, J., Poline, J.B., Rokem, A., Schaefer, G., Sochat, V., Triplett, W., Turner, J.A., Varoquaux, G., Poldrack, R.A. (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data, 3, 160044, doi:10.1038/sdata.2016.44.
· Jenkinson, M., Bannister, P., Brady, M., Smith, S. (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 17(2), 825-841.
· Li, X., Morgan, P.S., Ashburner, J., Smith, J., Rorden, C. (2016). The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods, 264, 47-56, doi:10.1016/j.jneumeth.2016.03.001.
· Tange, O. (2011). Gnu parallel-the command-line power tool. The USENIX Magazine, 36(1), 42-47.
· Winkler, A.M., Ridgway, G.R., Douaud, G., Nichols, T.E., Smith, S.M. (2016). Faster permutation inference in brain imaging. Neuroimage, 141, 502-516, doi:10.1016/j.neuroimage.2016.05.068.
· Yan, C.-G., Wang, X.-D., Lu, B. (2021). DPABISurf: data processing & analysis for brain imaging on surface. Sci Bull, 66(24), 2453-2455, doi: https://doi.org/10.1016/j.scib.2021.09.016.
· Yan, C.G., Wang, X.D., Zuo, X.N., Zang, Y.F. (2016). DPABI: Data Processing & Analysis for (Resting-State) Brain Imaging. Neuroinformatics, 14(3), 339-351, doi:10.1007/s12021-016-9299-4.
· Zang, Y., Jiang, T., Lu, Y., He, Y., Tian, L. (2004). Regional homogeneity approach to fMRI data analysis. Neuroimage, 22(1), 394-400, doi:http://dx.doi.org/10.1016/j.neuroimage.2003.12.030.
· Zang, Y.F., He, Y., Zhu, C.Z., Cao, Q.J., Sui, M.Q., Liang, M., Tian, L.X., Jiang, T.Z., Wang, Y.F. (2007). Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev, 29(2), 83-91, doi:10.1016/j.braindev.2006.07.002.
· Zou, Q.-H., Zhu, C.-Z., Yang, Y., Zuo, X.-N., Long, X.-Y., Cao, Q.-J., Wang, Y.-F., Zang, Y.-F. (2008). An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. Journal of Neuroscience Methods, 172(1), 137-141, doi:http://dx.doi.org/10.1016/j.jneumeth.2008.04.012.
· Zuo, X.-N., Xing, X.-X. (2014). Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: A systems neuroscience perspective. Neuroscience & Biobehavioral Reviews, 45, 100-118, doi:http://dx.doi.org/10.1016/j.neubiorev.2014.05.009.

DPABISurf pull Docker安装
老师您好,我使用linux的ubuntu系统,matlab2018b安装DPABISurf,已经安装好docker,但是进行pull DPABISurf Docker这一步,使用代码docker pull cgyan/dpabi时,很多文件下着下着就下不动了。
最后提示:error pulling image configuration: Get "https://production.cloudflare.docker.com/registry-v2/docker/registry/v2/blobs/sha256/13/13d6174a60b18e990091efd5e64edc9e5795ce8d708ea90f81a4f45ac060/data?verify=1636427975-UAQ6S2tcIHUpEX1pA%2FsOg7qaEy4%3D": dial tcp 104.18.123.25:443: i/o timeout
然后我看pull Docker可以load local file。所以我进到官网给的百度网盘上下载了dpabi60docker.tar.gz和gyancircosdocker.tar.gz,这两个文件,我直接load的话,会提示 [1BError processing tar file(exit status 1): setxattr("/usr/lib/node_modules/npm", trusted.overlay.opaque=y): operation not supported。
麻烦老师能解答一下,如何才能顺利在linux上安装好 DPABISurf。万分感谢。
这两个问题都没有遇到过,正常操作应该就可以
这两个问题都没有遇到过,正常操作应该就可以。可以试试换网络环境。
1. 换个网络 2. .tar.gz不要先解压缩
1. 换个网络
2. .tar.gz不要先解压缩
数据预处理出现错误
老师:
在使用dpabi进行数据运算时遇到如下错误,我使用的版本是matlab2017a,spm12,dpabi_V6.1。
怀疑是预处理出现问题,在论坛内搜索后又重新检查过文件夹,Check Data Organization也没有问题。再去检查数据的时候发现生成的FunImg文件夹中有几组数据缺失。(如下图)
当删去这些项目的数据再次使用dpabi预处理的时候又会发现之前没有问题的数据发生相同现象。故求助于老师,恳请老师指教,谢谢!
严老师您好!
严老师您好!
我是用dpabisurf处理结构像,无论我自己的数据还是您提供的demo都报错了。安装完后(window系统下)进行volume-surface project测试是能生成两个gii文件的。dapbi也升级到v6.1版本的,docker也是最新版的(没有配置处理器、内存跟工作文件夹的选项)。报错如下,烦请严老师帮忙解答,谢谢您!!
请下载最新的spm并在matlab中添加路径。
请下载最新的spm并在matlab中添加路径。
Some errors occur in using ADNI data
Hi
I am trying to use ADNI data in FC calculation.
I use template named "Calculate in MNI Space (warp by DARTEL)" in DPABI,
and in the RS-fMRI preprocessing, I only change values of "Parallel Workers : 8" and "Define ROI as AAL template".
When I try to use only one subject, it works well.
However, even though the preprocessing for each of single subject works well, whenever I try to use multiple subjects at once,
always an error occurs.
This is what I get.
As you are using Multi-Band,
As you are using Multi-Band, you can either skip slice timing correction.
Or setup very carefully according to http://rfmri.org/slicetiming
bug
老师您好!我在使用您的演示数据进行视频的展示时,出现了报错,请问是什么原因呢?如何解决呢?
'norminv’需要 Statistics and Machine Learning Toolbox。
刚刚接触,关于DPABISurf Pipeline设置中两个小问题!
1,数据DICOM格式,我这样设置slice number 0, slicer order [0], Reference slice 0,您教程中说这这里可以填写0,软件自动读取,对不对?slicer order 这里也是[0]?,还是按默认的[1 3 5 7 9...33 2 46 ...32]
2-CPU8核心,内存32G, 跑一个被试,您视频教程说Parrallel works 设置成1;如果一次跑4个被试,Parrallel works 填4就够了, 还是填8会快一些呢?
1. 对。都可以,不影响。 2. 填4就够了。
1. 对。都可以,不影响。
2. 填4就够了。
老师您好,我在进行数据预处理时,产生了下面两个问题:
老师您好,我在进行数据预处理时,产生了下面两个问题:
1、我使用的是EPI模板进行空间标准化的,协变量放在标准化之前和之后都会产生下面问题,这个问题对最终结果的影响大吗?
警告: 矩阵接近奇异值,或者缩放错误。结果可能不准确。RCOND = 1.724427e-09。
> In y_regress_ss (line 39)
In y_RegressOutImgCovariates (line 140)
2、我看老师当时的视频说要是采用EPI标准化要在标准化之后进行协变量回归,但我在查资料的时候有些也在说也可以先进行回归,我把两种方式都试了一遍,最后利用时间序列去分类时两种方法的准确率相差较大,想问一下老师,协变量究竟是放在标准化之前还是之后,放在之前和之后有什么影响呢?
老师,我想请问以下,为什么dpabi
老师,我想请问以下,为什么dpabi viewer里显著的cluster,map到brain net viewer里就不显示了呀
老师,我想请问以下,为什么dpabi
老师,我想请问以下,为什么dpabi viewer里显著的cluster,map到brain net viewer里就不显示了呀
严老师您好,我在进行dpabi预处理的时候
严老师您好,我在进行dpabi预处理的时候,进行到产生automask这一步会报出错误:(好像在第三个被试这里出了问题)
下标索引必须为正整数类型或逻辑类型。
出错 w_ClipLevel (line 50)
artifact removal
Thanks for developing dpabi/dparsfa!
Is reintroduction of noise during sequential preprocessing steps avoided in dparsfa? If not, is there going to be a fix for this? Linquist et al Hum Brain Mapp. 2019 Jun 1; 40(8): 2358–2376. doi: 10.1002/hbm.24528
老师好,跑完dpabisurf,ResultsS…
老师好,跑完dpabisurf,ResultsS/AnatSurfLH/fsaverage/Area里没有sz开头的结果,直接以ssub-sub*_space-fsaverage_hemi-L.area.gii去做统计吗,还是说需要做一下standardization,再比较
我的经验的话还是要先做z…
我的经验的话还是要先做z-standardization之后再统计比较好。