Dear Dr.YAN,
I am sorry to ask you a stupid question. Recently I want use the Rest to do seed-based FC, the input parameter contains some subjects(including patients and controls) with nii format, then voxel wise defined the roi, do all, fisher Z. The result only have two images in nii format which named FCMap and zFCMap, I don't think it can be used for later on two-sample t test. What's the wrong step do I made? And I also would like to know if I ues DPARSFA, is it will be a little difficult?
Best
Fish
Forums

You have 1 zFCMap for one
You have 1 zFCMap for one subject. And you will have a group of zFCMaps for a group of subjects. You can perform group analysis based on that.
Please see more details at http://rfmri.org/Course
DPARSFA would be easier even.
Best,
Chao-Gan
Dear Dr.Yan,
Dear Dr.Yan,
Thanks for your answers. Then I used dparsfa to do seed based FC, to be more precise, I based on the preprocessing data --FunImgRCWSDF to do FC, the seed is based on the result of ALFF after two-sample and binarized as a mask, the result of FC after two-sample t test is really strange, because it doesn't have any significant value. Do you thinks it is normal.
I am looking forward to your replies.
Best
Jing
Sometimes it's common that
Sometimes it's common that there were no significant differences between the two groups.
Best,
Chao-Gan
if the one-sample t test is right
Dear Dr.Yan,
Thanks so much for your replies. Here is the one-sample t-test of a seed-based FC, I mean the roi is based on the result of alff, but it looks a little weird, especially the result from spm in terms of whole clusters, it seems the cluster-level showed by spm only contains the whole brain. Do you think it is normal or wrong?
I am looking forward to your replies.
Best
Fish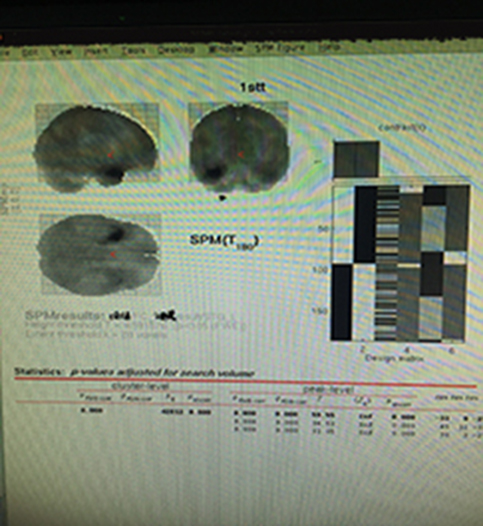
Hi,
Hi,
If you didn't perform global signal regression, the whole brain is highly correlated. You could raise the threshold.
Best,
Chao-Gan