Data Processing Assistant for Resting-State fMRI (DPARSF) V2.2
 |
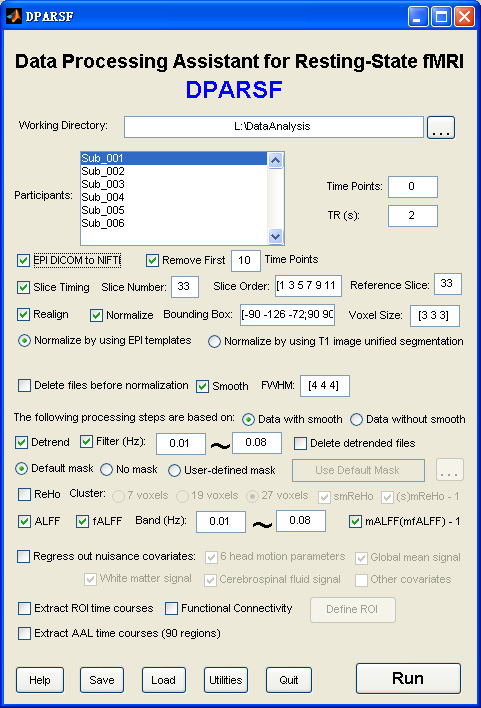
Data Processing Assistant for Resting-State fMRI (DPARSF) is a convenient plug-in software based on SPM and REST. You just need to arrange your DICOM files, and click a few buttons to set parameters, DPARSF will then give all the preprocessed (slice timing, realign, normalize, smooth) data, functional connectivity, ReHo, ALFF/fALFF, degree centrality, voxel-mirrored homotopic connectivity (VMHC) results. DPARSF can also create a report for excluding subjects with excessive head motion and generate a set of pictures for easily checking the effect of normalization. You can use DPARSF to extract ROI time courses efficiently if you want to perform small-world analysis. DPARSF basic edition is very easy to use while DPARSF advanced edition (alias: DPARSFA) is much more flexible and powerful. DPARSFA can parallel the computation for each subject, and can be used to reorient your images interactively or define regions of interest interactively. You can skip or combine the processing steps in DPARSF advanced edition freely. Please download a MULTIMEDIA COURSE to know more about how to use this software. Add DPARSF's directory to MATLAB's path and enter "DPARSF" or "DPARSFA" in the command window to enjoy DPARSF basic edition or advanced edition. The latest release is DPARSF_V2.2_130309.DOWNLOAD Multimedia Course: Data Processing of Resting-State fMRI |
1. Advanced Edition: Fixed a bug in "Smooth by DARTEL" caused in previous revision (DPARSF_V2.2_130214). (Thanks for the report of Maki Koyama).
New features of DPARSF_V2.2_130303:
1. Advanced Edition: Fixed a bug in processing of multiple sessions - only process the last session in normalization and smooth.
2. Basic Edition: Fixed a bug caused in the previous release (V2.2_130214) - cannot load the correct masks (Thanks for the report of Tao Yang).
New features of DPARSF_V2.2_130224:
1. This release fixed some minor bugs, but will not affect the results of any data analysis. The bugs appear in uncommon parameter settings and stop the processing in the worst cases.
2. Updated the DPARSFVersion information in the template parameter files.
3. DPARSF basic edition will also output the results of ReHo/ALFF/fALFF after Z-standardization (subtract the whole brain mean and divide by the whole brain standard deviation).
4. Fixed a bug in nuisance covariates regression when CovMat is not defined.
5. Fixed a bug that can not save the ROI signals correctly in text file if the data is not in double format. (Thanks for the report by H. Baetschmann)
6. Fixed a bug in creating mean functional image for 4D files. (Thanks for the report and revision by S. Orsolini)
7. Fixed a bug in displaying ROI Templates in linux system. (Thanks for the report by Han Zhang)
New features of DPARSF_V2.2_121225:
1. Support parallel computing! If you installed the MATLAB parallel computing toolbox, you can set the number of "Parallel Workers", DPARSFA will distribute the subjects into different CPU cores.
2. In addressing head motion concerns in resting-state fMRI analyses (Power et al., 2012; Satterthwaite et al., 2012; Van Dijk et al., 2012), we provide Friston 24-parameter correction (See Yan et al., 2013 for a comprehensive assessment of head motion on functional connectomics) as well as voxel-specific head motion calculation and correction (Satterthwaite et al., 2013; Yan et al., 2013). DPARSF also calculate the voxel-specific mean framewise displacement (FD) and volume-level mean FD (Power) (Power et al., 2012) or FD (Jenkinson) (i.e., relative RMS; Jenkinson et al., 2002) for accounting head motion at group-level analysis. The data scrubbing approach is also supported with different methods: 1) model each bad time point as a separate regressor in nuisance covariates regression, 2) delete bad time points, 3) interpolate bad time points with nearest neighbor, linear or cubic spline interpolation.
3. According to (Weissenbacher et al., 2009), the nuisance covariate regression could be performed before filtering and at very early stage. Users can also choose Template Parameters: TRADITIONAL order to have the same order as the previous version.
4. Support .nii.gz in all the steps. No longer need to convert 4D .nii.gz into 3D .img/.hdr. Simply put .nii.gz under FunImg or any starting directory name, DPARSFA will handle the .nii.gz by itself.
5. If the Number of Time Points is set to 0, then DPARSFA will not check the number of time points.
6. If TR is set to 0, then DPARSFA will retrieve the TR information from the NIfTI images. Please ensure the TR information in NIfTI images are correct!
7. If Slice Number is set to 0, then retrieve the slice number from the NIfTI images. The slice order is then assumed as interleaved scanning: [1:2:SliceNumber, 2:2:SliceNumber]. The reference slice is set to the slice acquired at the middle time point, i.e., ceil(SliceNumber/2). SHOULD BE EXTREMELY CAUTIOUS!!!
8. Support calculating resting-state fMRI metrics in native space and warping by DARTEL. The mask files and/or region of interest (ROI) files in standard space are warped into native space by using the parameters estimated in segmentation or DARTEL.
9. Spatial normalization and smooth can be performed on the calculated resting-state fMRI derivatives.
10. Supporting extracting ROI time course using masks with multiple labels. More template ROI definitions are supported: Dosenbach’s 160 functional ROIs (Dosenbach et al., 2010), Andrews-Hanna’s default mode network ROIs (Andrews-Hanna et al., 2010), Craddock’s clustering ROIs (Craddock et al., 2011), AAL atlas and Harvard-Oxford atlas.
11. More resting-state fMRI metrics are included, e.g., voxel-mirrored homotopic connectivity (VMHC) (Zuo et al., 2010), Degree Centrality (Buckner et al., 2009) and connectome-wide association studies based on multivariate distance matrix regression (Shehzad et al., 2011).
12. For VMHC analyses, support normalizing the data further to a symmetric template. 1) Get the T1 images in MNI space (e.g., wco*.img or wco*.nii under T1ImgNewSegment or T1ImgSegment) for each subject, and then create a mean T1 image template (averaged across all the subjects). 2) Create a symmetric T1 template by averaging the mean T1 template (created in Step 1) with it's flipped version (flipped over x axis). 3) Normalize the T1 image in MNI space (e.g., wco*.img or wco*.nii under T1ImgNewSegment or T1ImgSegment) for each subject to the symmetric T1 template (created in Step 2), and apply the transformations to the functional data (which have been normalized to MNI space beforehand). Please see a reference from Zuo et al., 2010.
13. A group analyses function was added as y_GroupAnalysis_Image by scripting call. T tests or F tests could be performed for a given set of regressors.
14. Template Parameters:
Calculate in Original Space (warp by DARTEL)
Calculate in Original Space (warp by information from unified segmentation)
Calculate in MNI Space (warp by DARTEL)
Calculate in MNI Space (warp by information from unified segmentation)
Calculate in MNI Space: TRADITIONAL order [This is the order used in DPARSF basie edition as well as DPARSFA V2.1]
Intraoperative Processing
Task fMRI data preprocessing
VBM (New Segment and DARTEL)
VBM (unified segmentation)
Blank
Many thanks to Dr. Chris Rorden for suggesting features 4-7. Many thanks to Dr. Susan Whitfield-Gabrieli for discussing the head motion scrubbing regressors.