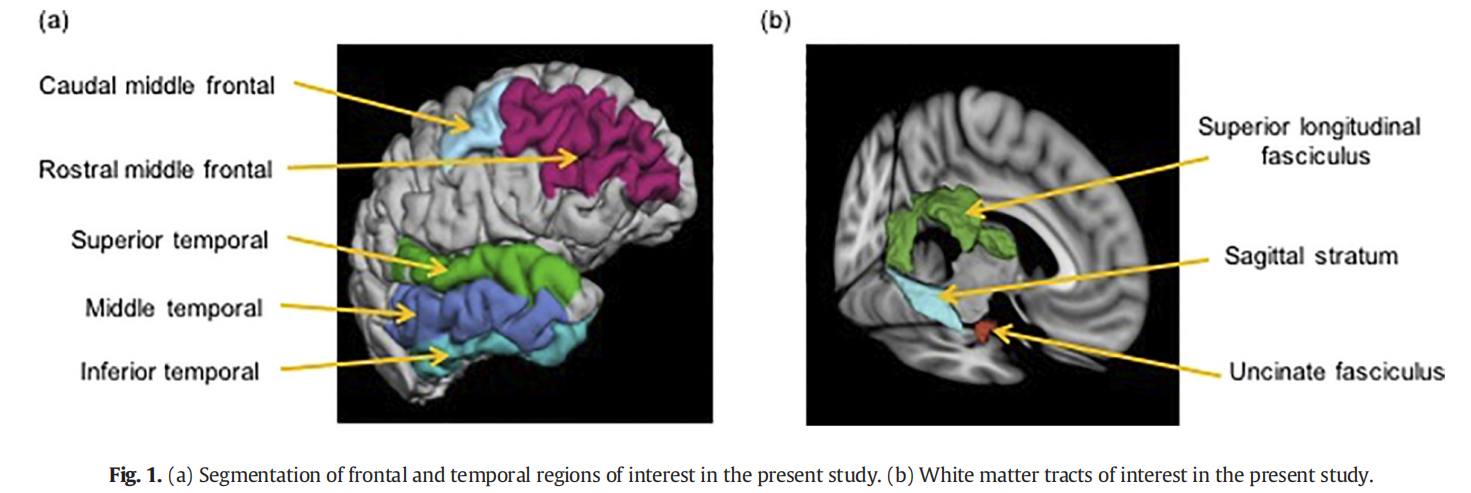
How to remove the edge of a cluster in the BrainNet Viewer
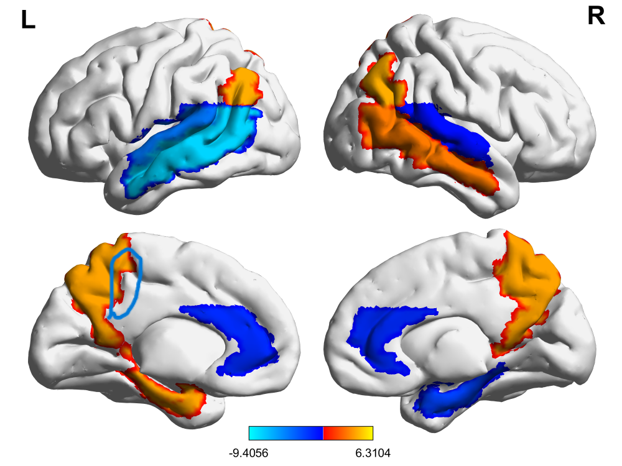
I found the BrainNet Viewer somtimes renders an odd "edge" of a cluster, as shown in the above picture. Especially when you wish to render a region of interest that consists of the same value for all voxels. You may change the default setting to remove it.
In the setting window, got to "Global" and change the "shading properties" as "Flat".