REST版本号V1.8_130615 DPARSFA版本号V2.3_130615 spm版本为spm8 matlab为linux下2010a(我测试的操作系统分别为redhat el5.7和redhat el6.2 测试用户分别为root和普通用户,普通用户有rest和dparsf文件夹下的777权限) 我执行dicom文件转nifti时遇到下面的情况FunRaw转FunImg时,FunImg文件夹会产生,但是nifti格式文件不会产生,程序会一直等待,直到我ctrl+c终止。如果用后面的脚本可以解决这一问题
这个问题我用fedora 14和opensuse 11.4测试过,同样版本的spm,matlab,REST和DPARSFA是可以转换的。但是我在虚拟机内安装的redhat el5.7就转换失败。请问是什么原因造成的?有没有可能是操作系统的内核问题?
不知道我上面的问题是否描述清楚了,如果您还需要其他信息的话我再进行补充?
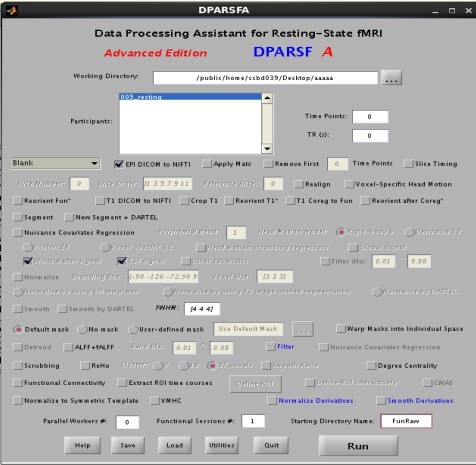
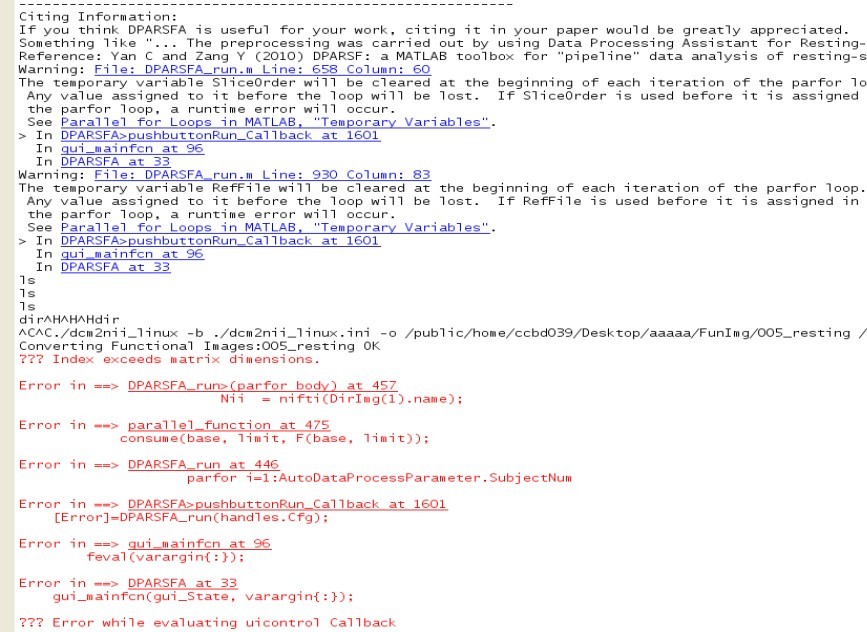
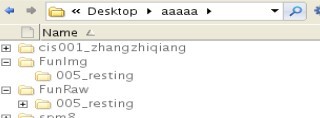
我看了y_Call_dcm2nii.m文件
OldDirTemp=pwd;
cd([ProgramPath,filesep,'dcm2nii']);
if ispc
if strcmpi(Option,'DefaultINI')
Option='-b dcm2nii.ini';
end
eval(['!dcm2nii.exe ',Option,' -o ',OutputDir,' ',InputFilename]);
elseif ismac
if strcmpi(Option,'DefaultINI')
Option='-b ./dcm2nii_linux.ini';
end
eval(['!./dcm2nii_mac ',Option,' -o ',OutputDir,' ',InputFilename]);
else
if strcmpi(Option,'DefaultINI')
Option='-b ./dcm2nii_linux.ini';
end
eval(['!chmod +x dcm2nii_linux']);
eval(['!./dcm2nii_linux ',Option,' -o ',OutputDir,' ',InputFilename]);
end
cd(OldDirTemp);
应该执行的是最后的一个else语句
用脚本可以转换文件格式
#!/bin/bash
rootdir="/home/***/frequency_ybk/analysis";
data_type="FunRaw";
for condition in {"EO","EC"};do
subname=048
if [ $data_type = "T1Raw" ];then
data_type_dst="T1Imgtmp";
elif [ $data_type = "FunRaw" ];then
data_type_dst="FunImgtmp";
elif [ $data_type = "3DT1Raw" ];then
data_type_dst="3DT1Imgtmp";
fi
echo $data_type
outrootdir="${rootdir}/${condition}/${data_type_dst}";
mkdir $outrootdir;
outdir="${outrootdir}/sub${subname}";
mkdir $outdir
datadir="${rootdir}/${condition}/${data_type}/sub${subname}";
cd $datadir;
fn="${datadir}/*.1"
cd /***/DPARSF_V2.2_130309/dcm2nii;
dcm2nii -b dcm2nii.ini -o ${outdir} ${fn};
done

"Calling other commands from MATLAB" choke the processing
Hi,
Thanks for your advice, the
Thanks for your advice, the problem has been resolved
too many .nii.gz files
Hi
I have been using DPARSF A for a while now. For some reason I am now getting the error message "too many nii.gz files in each subjects directory, should only keep one 4D .nii.gz files"
However I do not use 4D files, onle nii files. What could be the problem? Can anyone help?
Many thanks,
Stephanie
Re: [RFMRI]
回复:[RFMRI]