Hi, I wonder to know is there any manual \ppt\video for software GRETNA? I am a medical school student and going to analyze the patients' functional brain networks of small world properities with GRETNA. Manuals downloaded from the website of "NITRC" were too simple for me and i still could not know how to run it. It is difficult but i have to do this for my graduation. Besides that, i want to know is there any other software to analyze small world network for a beginner? Thank you very much! I will appreciate for your help!
Forums

Re: [RFMRI] GRETNA/Small
Dear Sandy Wang, I really
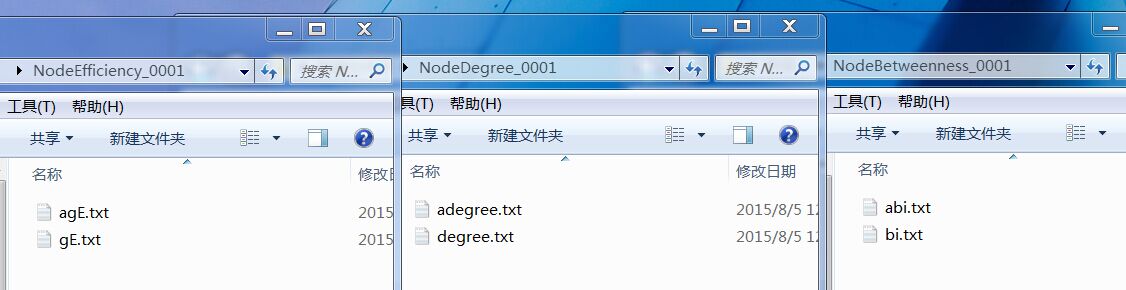
Dear Sandy Wang, I really appreciate for your kindhearted help. Actually my data has been calculated by my cooperator (you can see an example followed by my reply ), what are the most two difficult problems for me now is that i want to compare the nodal degree\nodal efficiency\nodal betweenness between patients and normal people, and i need a picture to reflect those changing nodes for patients group. But there are two kinds of "txt"(eg: bi.txt and abi.txt), i want to know that which kind of txt should i compare? Because abi.txt shows to be a numberical matrix(90*36, sparsity was set between 0.05 and 0.4 with an intervel of 0.01 ), i couldn't analyze those data by SPSS, i wonder to know could statistical tool in the GRETNA can help me to achive it? Could i add those calculated data into GRETNA and analyze them automaticly? If possible, there are only six statistical methods and which kind of statistical method i should use ? Thank you very much for your help! Looking forward to your reply!