Hi,
are the AllResampled_WhiteMask_09_91x109x91.nii files used for nuisance regression of white matter signal or the FunSpace_ThrdMask_subject_WM.nii files in the segmentation mask folder?I was wondering if it is somehow possible to change the mask that is used for the nuisance regression of white matter before regressing out covariates? In other words, since I'm interested in signals from brainstem I need to make sure that signals from this region are not regressed out as a white matter nuisance covariate. But the white matter mask that is created from the segmentation of SPM normally includes brainstem (specifically, the FunSpace_ThrdMask does but the AllResampled mask does not somehow).
many thanks in advance
Forums

Hi, please post a snapshot of
Hi, please post a snapshot of your parameter setting (snapshot of "Nuisance Regression Setting" is also needed). It depends on your setting to select the mask.
Thanks for the reply! I
Thanks for the reply!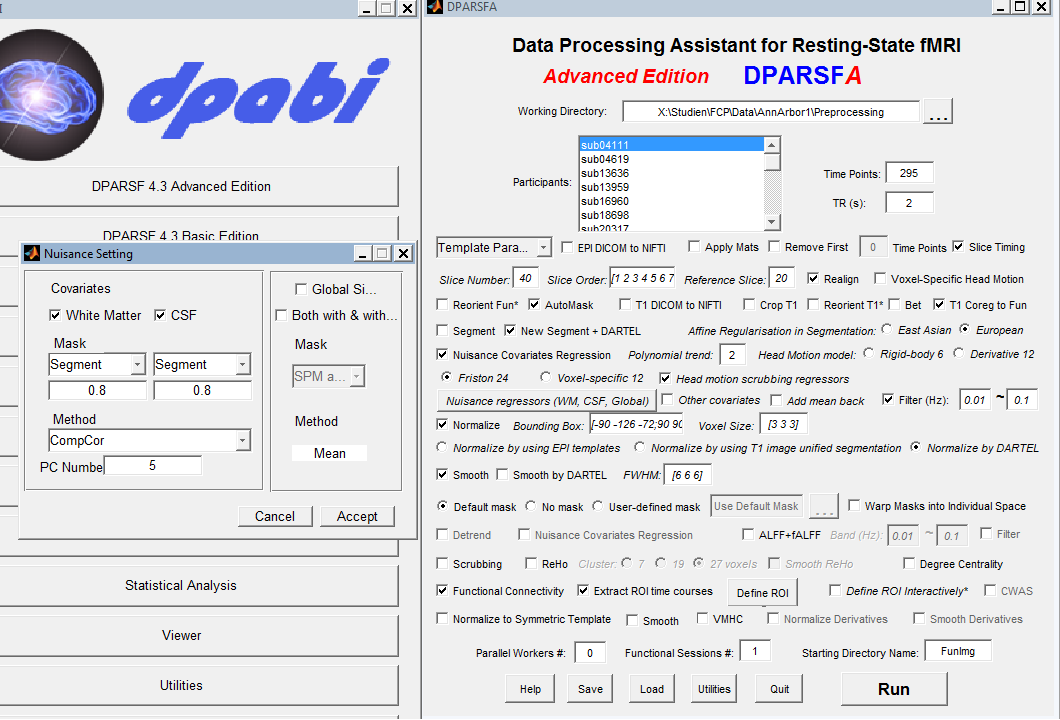 I attached a screenshot of my settings.
I attached a screenshot of my settings.
Hi David,
Hi David,
In your settings, these masks were used:
/Masks/SegmentationMasks/FunSpace_ThrdMask_Sub_001_WM.nii
/Masks/SegmentationMasks/FunSpace_ThrdMask_Sub_001_CSF.nii
Please consider to revise them.
Best,
Chao-Gan
Thanks for the reply,
Thanks for the reply,
I'm not sure how to do this, automatically. In other words, I need to remove a specific ROI e.g. Brain-Stem from the Mask BEFORE it will be used in the covariate regression. So this needs to happen right after the masks have been created so it can be used as input to the covariate regression and ideally, this does not interrupt the automatic processing of the data.
This is the tricky part and I'm not sure if this is easily possible with DPARSF, but maybe you know a way to do this?
Hi,
Hi,
There is not an easy way to do this.
/Masks/SegmentationMasks/FunSpace_ThrdMask_Sub_001_WM.nii
You can edit the mask and continue the remain alaysis starting with Nuisance covarites regression.
Best,
Chao-Gan
Thanks! That is a good idea.
Thanks! That is a good idea. The ThrdMasks are not normalized though, so the problem is to somehow remove the normalized ROI (brainstem) from the ThrdMask, which I don't know how to do. Do you maybe have an idea? :)
You can set brainstem as an
You can set brainstem as an ROI doing FC analysis, and using the calculating in original space option. This will transform the masks from MNI space to original space. Then you can apply to the ThrdMask.
Awesome, thanks for the
Awesome, thanks for the support! May I ask how the transform from MNI back to original space is done? Is there a function I can use?
You can have a look at y
You can have a look at y_WarpBackByDARTEL.m
Great, thank you so much!
Great, thank you so much!