How to perform spectral DCM analysis
- Read more about How to perform spectral DCM analysis
- Log in or register to post comments
Conjunction analysis
- Read more about Conjunction analysis
- 3 comments
- Log in or register to post comments
Dear Chao-Gan,
I have examined ALFF abnormalities in two type of patients and I would like to examine the shared abnormalities. Acctually, I have tried to use SPM to do a conjuntion analysis, but SPM seemed to require the spm.mat file which I did have, so I was wondering that can I achieve this goal using DPABI?
Best,
Vincent
Motion regressors - temporal derivatives
- Read more about Motion regressors - temporal derivatives
- 1 comment
- Log in or register to post comments
Dear Dr. Yan,
first of all, thank you for the wonderful tools you have created, which provide a great service to our community.
How to view the results of F.nii after ancova analysis
Hi,
Sorry to bother you again. I performed ANCOVA analysis with dpabi and obtained 10 nii images including the F.nii map. But I cannot view the results of F.nii using dpabi viewer. Some parameters of the F.nii can be viewed in dpabi viewer overlay list (see below).
Error in DPARSFA
- Read more about Error in DPARSFA
- 2 comments
- Log in or register to post comments
Hi, we are using your toolbox DPARSFA (v4.2) with matlab R2014b
We followed the tutorial concerning the folder structure.
We selected this parameters :
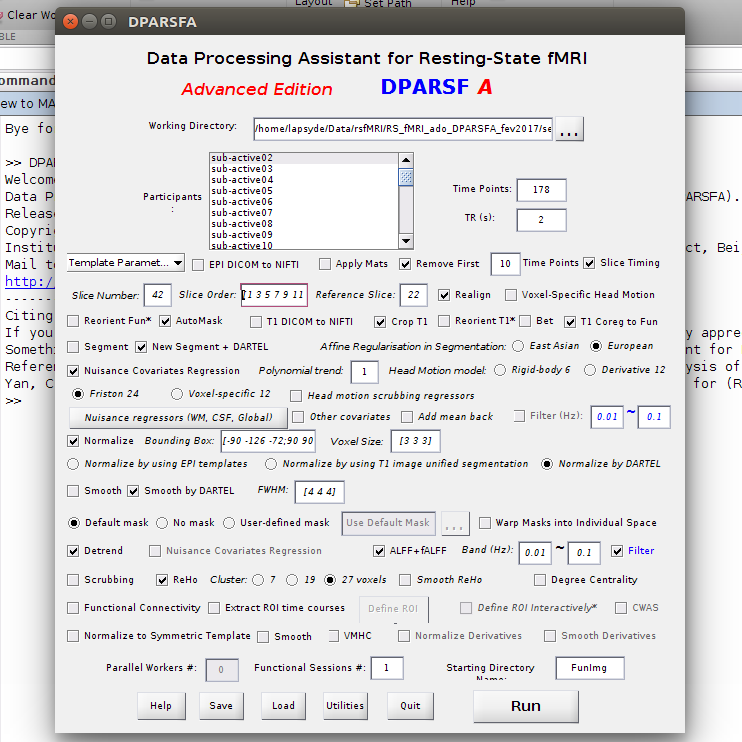
When we compute the data, we have the following error message :
Error with DPARSF in Matlab 2016a
- Read more about Error with DPARSF in Matlab 2016a
- 7 comments
- Log in or register to post comments
Hello,
Thank you for making these tools available to the public. I'm receiving the following error when I run DPARSF (latest version) in Matlab 2016a:
Error using Save subjects list in QC
- Read more about Error using Save subjects list in QC
- 1 comment
- Log in or register to post comments
Hi Dr. Yan,
When I used the "Save subjects list" in QC, there was a error:
How to add only ROI when I I finished the whole analysis?
Dear Chao-Gan,
I use the spm8, MatlabR2014b and DPSRASF basic 2.3_130615.
My analysis lasts 10 h for 20 patients. I would like add only next ROI. My question is, who I have to choose a folder ? And sekend: do I have of defining "time point" again? I have mistake and dparsf not responding. I've tried different ways.
Thanks in advance for your help.
Regards,
Ilona
How to use the results of ANCOVA and make statistical inference?
Hi Yan,
I perfomed ANCOVA and chose tukey as post hoc analysis to find the diffefence of 3 groups, however I don't know how to explain the results.
An error occured when I tried to view the results of F.nii using dpabi viewer.
Error DPABI viewer under Matlab 2016a
- Read more about Error DPABI viewer under Matlab 2016a
- 1 comment
- Log in or register to post comments
Hi,
It seems that I can't run DPABI properly under Matlab 2016a. When I start dpabi I get the following error and the logo doesn't show up (but dpabi starts without showing the logo):
