multiband 数据预处理错误
- Read more about multiband 数据预处理错误
- 5 comments
- Log in or register to post comments
老师:
您好!
我是DPABI的初学者,我用的是DPABI3.1,matlab2014b,spm12.
想请教几个问题:
1、在设置time points和TR为0后得到的TRInfo.tsv文件打开得到的time points 是1,但实际time points是150,请问为什么会出现这种情况?
2、在转dicom文件为nifti之后进行其他处理时,无论设置time points为1还是150,都会出现如下错误
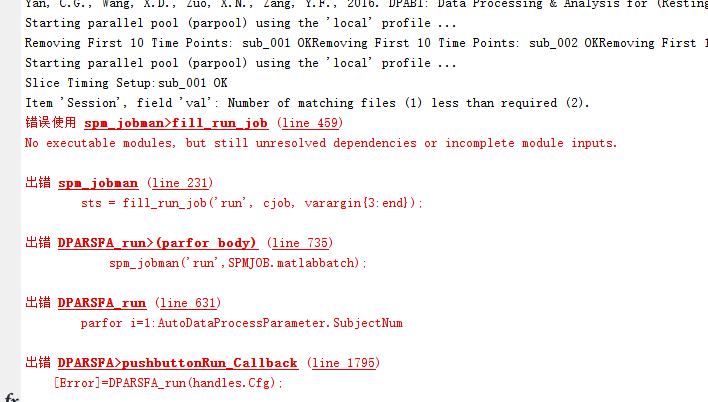
请问该如何解决?
3、multiband数据如何设置slice number和slice order以及reference slice?
