DPABIFiber is a fiber tractography analysis toolbox based on diffusion-weighted imaging (DWI), evolved from DPABI/DPABISurf/DPABINet/DPARSF, as easy-to-use as DPABI/DPABISurf/DPABINet/DPARSF. DPABIFiber is based on QSIPrep (Cieslak et al., 2021), MRtrix3 (Tournier et al., 2019), AFQ (Yeatman et al., 2012), fMRIPprep (Esteban et al., 2019), FreeSurfer (Tustison et al., 2014), ANTs (Avants et al., 2009), FSL (Jenkinson et al., 2012), SPM12 (Ashburner, 2012), dcm2niix (Li et al., 2016), PALM (Winkler et al., 2014), GNU Parallel (Tange, 2011), MATLAB (The MathWorks Inc., Natick, MA, US), Docker (https://docker.com) and DPABI (Yan et al., 2016). DPABIFiber provides a user-friendly graphical user interface (GUI) for pipeline DWI preprocessing, fiber tractography reconstruction, tract-based spatial statistics (TBSS) (Smith et al., 2006), automating fiber-tract quantification (AFQ) (Yeatman et al., 2012), structural connectome matrix analyses, seed-based structural connectivity analyses, and tract-weighted functional connectivity (TW-FC) (Calamante et al., 2013), while requires no programming/scripting skills from the users.
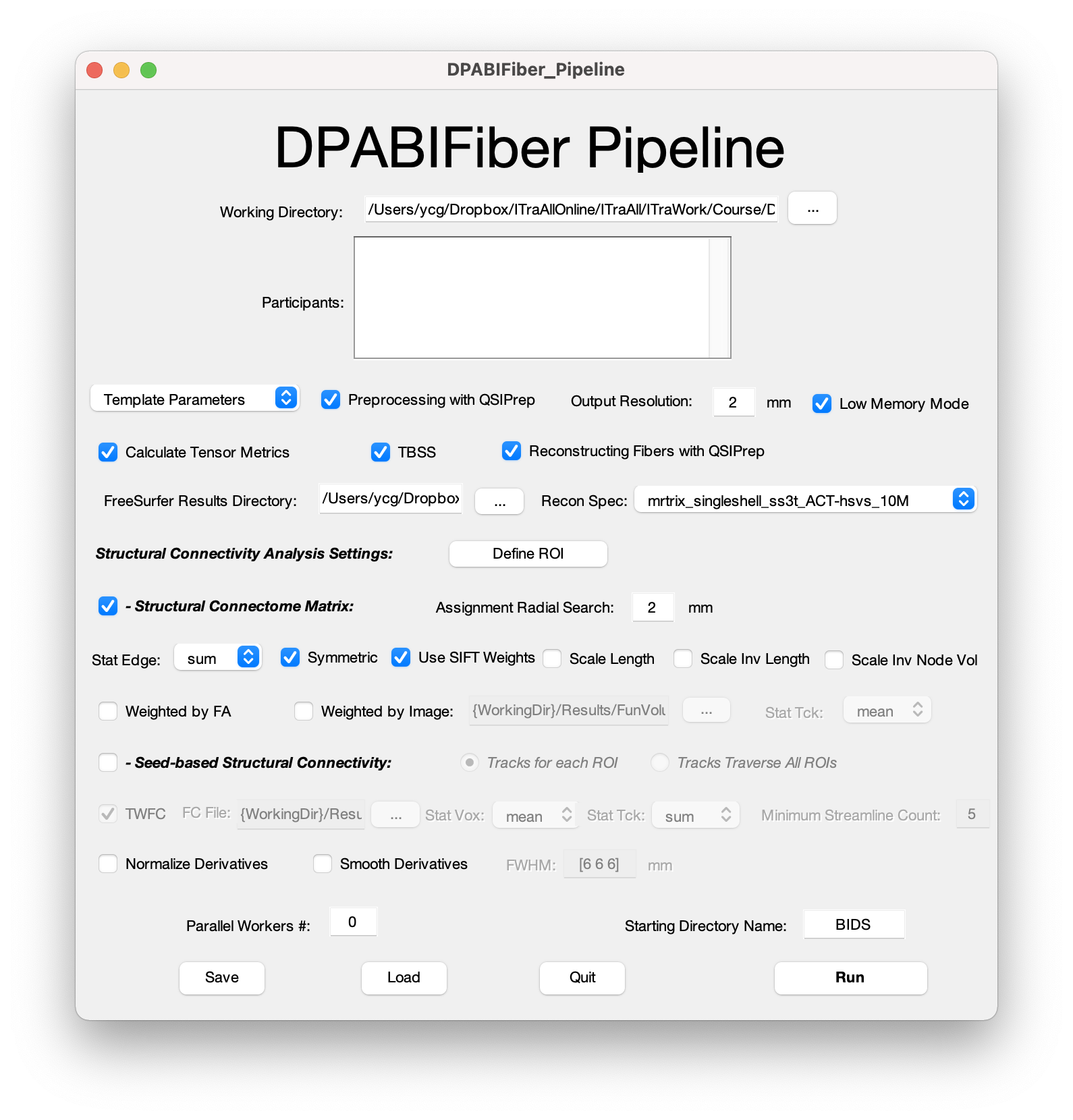
The DPABIFiber pipeline first preprocesses the raw T1 and DWI data with QSIPrep, which integrates FSL and ANTs. QSIPrep preprocesses data by conforming image and gradient orientation, grouping by distortion, denoising, distortion correction, head motion correction, building b0 templates, spatial registration and normalization. Then, the tensors and the DWI metrics including apparent diffusion coefficient (ADC), fractional anisotropy (FA), axial diffusivity (AD), and radial diffusivity (RD) are derived from preprocessed DWI data using MRtrix3 and subsequently generate TBSS results using FSL. After that, the QSIPrep is called again to reconstruct fiber tracts. In this step, DPABIFiber adopts two sets of optimized fiber reconstruction workflows, including anatomically-constrained tractography (Smith, Tournier et al. 2012) (ACT) with hybrid surface volume segmentation (Smith, Skoch et al. 2020) (hsvs) and AFQ. Both single-shell acquisition and multi-shell acquisition images are compatible. Based on the constructed fiber tract, advanced analysis such as structural connectome matrix and seed-based structural connectivity with predefined regions of interest (ROI) could be further conducted. While generating structural connectome matrices, multiple edge defining options (e.g. sum, max and mean) and weighting options (e.g. weight by edge length, inverted edge length and inverted node volume) were available for various research purposes. For seed-based structural connectivity analysis, users can choose to generate tracks for each ROI or traverse all ROIs, and calculate TW-FC with abundant optional settings. Finally, derivatives are normalized and smoothed.
DPABIFiber is designed to make fiber tractography analysis require minimum manual operations and almost no programming/scripting experience. We anticipate this updated open-source toolbox will assist novices and expert users alike and continue to support advancing structural MRI methodology and its application to clinical translational studies.
References:
Ashburner, J. (2012). SPM: a history. NeuroImage, 62(2), 791-800.
Avants, B.B., Tustison, N., Song, G. (2009). Advanced normalization tools (ANTS). Insight j, 2(365), 1-35.
Calamante, F., Masterton, R.A., Tournier, J.D., Smith, R.E., Willats, L., Raffelt, D., Connelly, A. (2013). Track-weighted functional connectivity (TW-FC): a tool for characterizing the structural-functional connections in the brain. Neuroimage, 70, 199-210, doi:10.1016/j.neuroimage.2012.12.054.
Cieslak, M., Cook, P.A., He, X., Yeh, F.C., Dhollander, T., Adebimpe, A., Aguirre, G.K., Bassett, D.S., Betzel, R.F., Bourque, J., Cabral, L.M., Davatzikos, C., Detre, J.A., Earl, E., Elliott, M.A., Fadnavis, S., Fair, D.A., Foran, W., Fotiadis, P., Garyfallidis, E., Giesbrecht, B., Gur, R.C., Gur, R.E., Kelz, M.B., Keshavan, A., Larsen, B.S., Luna, B., Mackey, A.P., Milham, M.P., Oathes, D.J., Perrone, A., Pines, A.R., Roalf, D.R., Richie-Halford, A., Rokem, A., Sydnor, V.J., Tapera, T.M., Tooley, U.A., Vettel, J.M., Yeatman, J.D., Grafton, S.T., Satterthwaite, T.D. (2021). QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat Methods, 18(7), 775-778, doi:10.1038/s41592-021-01185-5.
Esteban, O., Markiewicz, C.J., Blair, R.W., Moodie, C.A., Isik, A.I., Erramuzpe, A., Kent, J.D., Goncalves, M., DuPre, E., Snyder, M., Oya, H., Ghosh, S.S., Wright, J., Durnez, J., Poldrack, R.A., Gorgolewski, K.J. (2019). fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature Medicine, 16(1), 111-116, doi:10.1038/s41592-018-0235-4.
Jenkinson, M., Beckmann, C.F., Behrens, T.E., Woolrich, M.W., Smith, S.M. (2012). Fsl. NeuroImage, 62(2), 782-790.
Li, X., Morgan, P.S., Ashburner, J., Smith, J., Rorden, C. (2016). The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods, 264, 47-56, doi:10.1016/j.jneumeth.2016.03.001.
Smith, R., Skoch, A., Bajada, C.J., Caspers, S., Connelly, A. (2020). Hybrid surface-volume segmentation for improved anatomically-constrained tractography.
Smith, R.E., Tournier, J.D., Calamante, F., Connelly, A. (2012). Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. NeuroImage, 62(3), 1924-1938, doi:10.1016/j.neuroimage.2012.06.005.
Smith, S.M., Jenkinson, M., Johansen-Berg, H., Rueckert, D., Nichols, T.E., Mackay, C.E., Watkins, K.E., Ciccarelli, O., Cader, M.Z., Matthews, P.M., Behrens, T.E. (2006). Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage, 31(4), 1487-1505, doi:10.1016/j.neuroimage.2006.02.024.
Tange, O. (2011). Gnu parallel-the command-line power tool. The USENIX Magazine, 36(1), 42-47.
Tournier, J.D., Smith, R., Raffelt, D., Tabbara, R., Dhollander, T., Pietsch, M., Christiaens, D., Jeurissen, B., Yeh, C.H., Connelly, A. (2019). MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage, 202, 116137, doi:10.1016/j.neuroimage.2019.116137.
Tustison, N.J., Cook, P.A., Klein, A., Song, G., Das, S.R., Duda, J.T., Kandel, B.M., van Strien, N., Stone, J.R., Gee, J.C., Avants, B.B. (2014). Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. NeuroImage, 99, 166-179, doi:10.1016/j.neuroimage.2014.05.044.
Winkler, A.M., Ridgway, G.R., Webster, M.A., Smith, S.M., Nichols, T.E. (2014). Permutation inference for the general linear model. NeuroImage, 92, 381-397, doi:10.1016/j.neuroimage.2014.01.060.
Yan, C.-G., Wang, X.-D., Zuo, X.-N., Zang, Y.-F. (2016). DPABI: data processing & analysis for (resting-state) brain imaging. Neuroinformatics, 14(3), 339-351.
Yeatman, J.D., Dougherty, R.F., Myall, N.J., Wandell, B.A., Feldman, H.M. (2012). Tract profiles of white matter properties: automating fiber-tract quantification. PLoS ONE, 7(11), e49790, doi:10.1371/journal.pone.0049790.


DPABIFiber
教授,您好。我通过您的慕课学习完了DPABIFiber,但是目前有一个问题就是慕课中DPABIFiber的数据预处理需要DPABISurf,而且是需要静息态。但就我而言,我的数据是没有静息态的,我也尝试把T1和DWI转为BIDS,但是没法跑成功。所以我有一些疑惑,DPABIFiber可以跑没有静息态的数据吗,谢谢教授。
您可以下载公开数据中的功能像创建一个“假的…
您可以下载公开数据中的功能像创建一个“假的”FunRaw文件夹。